Physical interaction between components of DNA mismatch repair and nucleotide excision repair
- PMID: 9826691
- PMCID: PMC24364
- DOI: 10.1073/pnas.95.24.14278
Physical interaction between components of DNA mismatch repair and nucleotide excision repair
Abstract
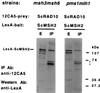
Nucleotide excision repair (NER) and DNA mismatch repair are required for some common processes although the biochemical basis for this requirement is unknown. Saccharomyces cerevisiae RAD14 was identified in a two-hybrid screen using MSH2 as "bait," and pairwise interactions between MSH2 and RAD1, RAD2, RAD3, RAD10, RAD14, and RAD25 subsequently were demonstrated by two-hybrid analysis. MSH2 coimmunoprecipitated specifically with epitope-tagged versions of RAD2, RAD10, RAD14, and RAD25. MSH2 and RAD10 were found to interact in msh3 msh6 and mlh1 pms1 double mutants, suggesting a direct interaction with MSH2. Mutations in MSH2 increased the UV sensitivity of NER-deficient yeast strains, and msh2 mutations were epistatic to the mutator phenotype observed in NER-deficient strains. These data suggest that MSH2 and possibly other components of DNA mismatch repair exist in a complex with NER proteins, providing a biochemical and genetical basis for these proteins to function in common processes.
Figures





Similar articles
-
Coordination of Rad1-Rad10 interactions with Msh2-Msh3, Saw1 and RPA is essential for functional 3' non-homologous tail removal.Nucleic Acids Res. 2018 Jun 1;46(10):5075-5096. doi: 10.1093/nar/gky254. Nucleic Acids Res. 2018. PMID: 29660012 Free PMC article.
-
Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination.Proc Natl Acad Sci U S A. 1997 Aug 19;94(17):9214-9. doi: 10.1073/pnas.94.17.9214. Proc Natl Acad Sci U S A. 1997. PMID: 9256462 Free PMC article.
-
Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae.Genetics. 1996 Mar;142(3):727-36. doi: 10.1093/genetics/142.3.727. Genetics. 1996. PMID: 8849883 Free PMC article.
-
DNA binding properties of the yeast Msh2-Msh6 and Mlh1-Pms1 heterodimers.Biol Chem. 2002 Jun;383(6):969-75. doi: 10.1515/BC.2002.103. Biol Chem. 2002. PMID: 12222686 Review.
-
Nucleotide excision repair in yeast.Mutat Res. 2000 Jun 30;451(1-2):13-24. doi: 10.1016/s0027-5107(00)00037-3. Mutat Res. 2000. PMID: 10915862 Review.
Cited by
-
Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae.Genetics. 2010 Jan;184(1):27-42. doi: 10.1534/genetics.109.107482. Epub 2009 Oct 19. Genetics. 2010. PMID: 19841096 Free PMC article.
-
Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination.Mol Cell Biol. 1999 Nov;19(11):7558-67. doi: 10.1128/MCB.19.11.7558. Mol Cell Biol. 1999. PMID: 10523644 Free PMC article.
-
Targeting and processing of site-specific DNA interstrand crosslinks.Environ Mol Mutagen. 2010 Jul;51(6):527-39. doi: 10.1002/em.20557. Environ Mol Mutagen. 2010. PMID: 20196133 Free PMC article. Review.
-
Regulation of mitotic homeologous recombination in yeast. Functions of mismatch repair and nucleotide excision repair genes.Genetics. 2000 Jan;154(1):133-46. doi: 10.1093/genetics/154.1.133. Genetics. 2000. PMID: 10628975 Free PMC article.
-
Transcription-coupled repair of DNA damage: unanticipated players, unexpected complexities.Am J Hum Genet. 1999 May;64(5):1259-63. doi: 10.1086/302390. Am J Hum Genet. 1999. PMID: 10205254 Free PMC article. Review. No abstract available.
References
-
- Kolodner R. Genes Dev. 1996;10:1433–1442. - PubMed
-
- Drummond J T, Li G-M, Longley M J, Modrich P. Science. 1995;268:1909–1912. - PubMed
-
- Johnson R E, Kovvali G K, Prakash L, Prakash S. J Biol Chem. 1996;271:7285–7288. - PubMed
-
- Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

