Differential tropism and replication kinetics of human immunodeficiency virus type 1 isolates in thymocytes: coreceptor expression allows viral entry, but productive infection of distinct subsets is determined at the postentry level
- PMID: 9811677
- PMCID: PMC110433
- DOI: 10.1128/JVI.72.12.9441-9452.1998
Differential tropism and replication kinetics of human immunodeficiency virus type 1 isolates in thymocytes: coreceptor expression allows viral entry, but productive infection of distinct subsets is determined at the postentry level
Abstract
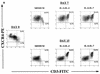
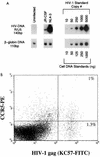
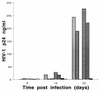
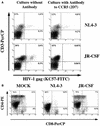
Human thymocytes are readily infected with human immunodeficiency virus type 1 (HIV-1) in vivo and in vitro. In this study, we found that the kinetics of replication and cytopathic effects of two molecular isolates, NL4-3 and JR-CSF, in postnatal thymocytes are best explained by the distribution of chemokine receptors used for viral entry. CXCR4 was expressed at high levels on most thymocytes, whereas CCR5 expression was restricted to only 0.1 to 2% of thymocytes. The difference in the amount of proviral DNA detected after infection of fresh thymocytes with NL4-3 or JR-CSF correlated with the levels of CXCR4 and CCR5 surface expression. Anti-CCR5 blocking studies showed that low levels of CCR5 were necessary and sufficient for JR-CSF entry in thymocytes. Interleukin-2 (IL-2), IL-4, and IL-7, cytokines normally present in the thymus, influenced the expression of CXCR4 and CCR5 on thymocytes and thus increased the infectivity and spread of both NL4-3 and JR-CSF in culture. NL4-3 was produced by both immature and mature thymocytes, whereas JR-CSF production was restricted to the mature CD1(-)/CD69(+) population. Although CXCR4 and CCR5 distribution readily explained viral entry in mature CD69(+) and immature CD69(-) cells, and correlated with proviral DNA distribution, we found that viral production was favored in CD69(+) cells. Therefore, while expression of CD4 and appropriate coreceptors are essential determinants of viral entry, factors related to activation and stage-specific maturation contribute to HIV-1 replication in thymocyte subsets. These results have direct implications for HIV-1 pathogenesis in pediatric patients.
Figures




Similar articles
-
CXCR4 and CCR5 on human thymocytes: biological function and role in HIV-1 infection.J Immunol. 1998 Sep 15;161(6):3103-13. J Immunol. 1998. PMID: 9743377
-
Differential tropism of HIV-1 isolates for distinct thymocyte subsets in vitro.AIDS. 1996 Jun;10(7):F9-16. doi: 10.1097/00002030-199606001-00001. AIDS. 1996. PMID: 8805858
-
Impact of cytokines on replication in the thymus of primary human immunodeficiency virus type 1 isolates from infants.J Virol. 2002 Jul;76(14):6929-43. doi: 10.1128/jvi.76.14.6929-6943.2002. J Virol. 2002. PMID: 12072494 Free PMC article.
-
Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease.Annu Rev Immunol. 1999;17:657-700. doi: 10.1146/annurev.immunol.17.1.657. Annu Rev Immunol. 1999. PMID: 10358771 Review.
-
HIV-1 coreceptors and their inhibitors.Curr Top Microbiol Immunol. 2006;303:97-120. doi: 10.1007/978-3-540-33397-5_5. Curr Top Microbiol Immunol. 2006. PMID: 16570858 Review.
Cited by
-
Exposure to human immunodeficiency virus/hepatitis C virus in hepatic and stellate cell lines reveals cooperative profibrotic transcriptional activation between viruses and cell types.Hepatology. 2016 Dec;64(6):1951-1968. doi: 10.1002/hep.28766. Epub 2016 Oct 5. Hepatology. 2016. PMID: 27531241 Free PMC article.
-
Histoarchitectural Deterioration of Lymphoid Tissues in HIV-1 Infection and in Aging.AIDS Res Hum Retroviruses. 2019 Nov/Dec;35(11-12):1148-1159. doi: 10.1089/AID.2019.0156. Epub 2019 Oct 7. AIDS Res Hum Retroviruses. 2019. PMID: 31474115 Free PMC article. Review.
-
Functional reconstitution of thymopoiesis after human immunodeficiency virus infection.J Virol. 2000 Mar;74(6):2943-8. doi: 10.1128/jvi.74.6.2943-2948.2000. J Virol. 2000. PMID: 10684316 Free PMC article.
-
Human immunodeficiency virus type 1 pathogenesis in SCID-hu mice correlates with syncytium-inducing phenotype and viral replication.J Virol. 2000 Apr;74(7):3196-204. doi: 10.1128/jvi.74.7.3196-3204.2000. J Virol. 2000. PMID: 10708436 Free PMC article.
-
Generation of SIV-resistant T cells and macrophages from nonhuman primate induced pluripotent stem cells with edited CCR5 locus.Stem Cell Reports. 2022 Apr 12;17(4):953-963. doi: 10.1016/j.stemcr.2022.03.003. Epub 2022 Mar 31. Stem Cell Reports. 2022. PMID: 35364011 Free PMC article.
References
-
- Aldrovandi G M, Feuer G, Gao L, Jamieson B, Kristeva M, Chen I S Y, Zack J A. The SCID-hu mouse as a model for HIV-1 infection. Nature. 1993;363:732–736. - PubMed
-
- Alkhatib G, Combardiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

