Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N
- PMID: 9789003
- PMCID: PMC23627
- DOI: 10.1073/pnas.95.22.12848
Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N
Abstract
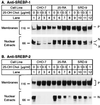
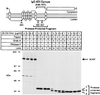
SREBP cleavage activating protein (SCAP), a membrane-bound glycoprotein, regulates the proteolytic activation of sterol regulatory element binding proteins (SREBPs), which are membrane-bound transcription factors that control lipid synthesis in animal cells. SCAP-stimulated proteolysis releases active fragments of SREBPs from membranes of the endoplasmic reticulum and allows them to enter the nucleus where they activate transcription. Sterols such as 25-hydroxycholesterol inactivate SCAP, suppressing SREBP proteolysis and turning off cholesterol synthesis. We here report the isolation of Chinese hamster ovary cells with a point mutation in SCAP (Y298C) that renders the protein resistant to inhibition by 25-hydroxycholesterol. Like the previously described D443N mutation, the Y298C mutation occurs within the putative sterol-sensing domain, which is part of the polytopic membrane attachment region of SCAP. Cells that express SCAP(Y298C) continued to process SREBPs in the presence of 25-hydroxycholesterol and hence they resisted killing by this sterol. In wild-type Chinese hamster ovary cells the N-linked carbohydrate chains of SCAP were mostly in the endoglycosidase H-sensitive form when cells were grown in medium containing 25-hydroxycholesterol. In contrast, when cells were grown in sterol-depleted medium, these chains were converted to an endoglycosidase H-resistant form. 25-Hydroxycholesterol had virtually no effect in cells expressing SCAP(D443N) or SCAP(Y298C). The relation between this regulated carbohydrate processing to the SCAP-regulated proteolysis of SREBP remains to be explored.
Figures
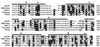


Similar articles
-
Three mutations in sterol-sensing domain of SCAP block interaction with insig and render SREBP cleavage insensitive to sterols.Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):16672-7. doi: 10.1073/pnas.262669399. Epub 2002 Dec 13. Proc Natl Acad Sci U S A. 2002. PMID: 12482938 Free PMC article.
-
Recurrent G-to-A substitution in a single codon of SREBP cleavage-activating protein causes sterol resistance in three mutant Chinese hamster ovary cell lines.Proc Natl Acad Sci U S A. 1996 Nov 26;93(24):13709-14. doi: 10.1073/pnas.93.24.13709. Proc Natl Acad Sci U S A. 1996. PMID: 8942999 Free PMC article.
-
Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein.Cell. 1996 Nov 1;87(3):415-26. doi: 10.1016/s0092-8674(00)81362-8. Cell. 1996. PMID: 8898195
-
Maintaining cholesterol homeostasis: sterol regulatory element-binding proteins.World J Gastroenterol. 2004 Nov 1;10(21):3081-7. doi: 10.3748/wjg.v10.i21.3081. World J Gastroenterol. 2004. PMID: 15457548 Free PMC article. Review.
-
Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes.Prog Lipid Res. 2001 Nov;40(6):439-52. doi: 10.1016/s0163-7827(01)00010-8. Prog Lipid Res. 2001. PMID: 11591434 Review.
Cited by
-
Juxtamembranous aspartic acid in Insig-1 and Insig-2 is required for cholesterol homeostasis.Proc Natl Acad Sci U S A. 2006 Apr 18;103(16):6154-9. doi: 10.1073/pnas.0601923103. Epub 2006 Apr 10. Proc Natl Acad Sci U S A. 2006. PMID: 16606821 Free PMC article.
-
Retrospective on Cholesterol Homeostasis: The Central Role of Scap.Annu Rev Biochem. 2018 Jun 20;87:783-807. doi: 10.1146/annurev-biochem-062917-011852. Epub 2017 Aug 25. Annu Rev Biochem. 2018. PMID: 28841344 Free PMC article. Review.
-
Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase.Crit Rev Biochem Mol Biol. 2010 Jun;45(3):185-98. doi: 10.3109/10409238.2010.485605. Crit Rev Biochem Mol Biol. 2010. PMID: 20482385 Free PMC article. Review.
-
Cholesterol and Its Derivatives: Multifaceted Players in Breast Cancer Progression.Front Oncol. 2022 May 26;12:906670. doi: 10.3389/fonc.2022.906670. eCollection 2022. Front Oncol. 2022. PMID: 35719918 Free PMC article.
-
Cholesterol feedback: from Schoenheimer's bottle to Scap's MELADL.J Lipid Res. 2009 Apr;50 Suppl(Suppl):S15-27. doi: 10.1194/jlr.R800054-JLR200. Epub 2008 Oct 29. J Lipid Res. 2009. PMID: 18974038 Free PMC article. Review.
References
-
- Brown M S, Goldstein J L. Cell. 1997;89:331–340. - PubMed
-
- Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. - PubMed
-
- Duncan E A, Brown M S, Goldstein J L, Sakai J. J Biol Chem. 1997;272:12778–12785. - PubMed
-
- Duncan E A, Dave U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. - PubMed
-
- Nohturfft A, Brown M S, Goldstein J L. J Biol Chem. 1998;273:17243–17250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

