Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage
- PMID: 9736709
- PMCID: PMC21615
- DOI: 10.1073/pnas.95.19.11175
Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage
Abstract
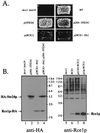
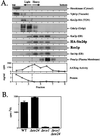
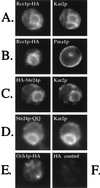
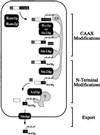
Proteins terminating in the CAAX motif, for example Ras and the yeast a-factor mating pheromone, are prenylated, trimmed of their last three amino acids, and carboxyl-methylated. The enzymes that mediate these activities, collectively referred to as CAAX processing components, have been identified genetically in Saccharomyces cerevisiae. Whereas the Ram1p/Ram2p prenyltransferase is a cytosolic soluble enzyme, sequence analysis predicts that the other CAAX processing components, the Rce1p and Ste24p proteases and the Ste14p methyltransferase, contain multiple membrane spans. To determine the intracellular site(s) at which CAAX processing occurs, we have examined the localization of the CAAX proteases Rce1p and Ste24p by subcellular fractionation and indirect immunofluorescence. We find that both of these proteases are associated with the endoplasmic reticulum (ER) membrane. In addition to having a role in CAAX processing, the Ste24p protease catalyzes the first of two cleavage steps that remove the amino-terminal extension from the a-factor precursor, suggesting that the first amino-terminal processing step of a-factor maturation also occurs at the ER membrane. The ER localization of Ste24p is consistent with the presence of a carboxyl-terminal dilysine ER retrieval motif, although we find that mutation of this motif does not result in mislocalization of Ste24p. Because the ER is not the ultimate destination for a-factor or most CAAX proteins, our results imply that a mechanism must exist for the intracellular routing of CAAX proteins from the ER membrane to other cellular sites.
Figures
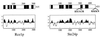
Similar articles
-
Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing.J Cell Biol. 1998 Aug 10;142(3):635-49. doi: 10.1083/jcb.142.3.635. J Cell Biol. 1998. PMID: 9700155 Free PMC article.
-
Proteolytic processing of certain CaaX motifs can occur in the absence of the Rce1p and Ste24p CaaX proteases.Yeast. 2009 Aug;26(8):451-63. doi: 10.1002/yea.1678. Yeast. 2009. PMID: 19504624 Free PMC article.
-
Saccharomyces cerevisiae a-factor mutants reveal residues critical for processing, activity, and export.Eukaryot Cell. 2006 Sep;5(9):1560-70. doi: 10.1128/EC.00161-06. Eukaryot Cell. 2006. PMID: 16963638 Free PMC article.
-
CaaX converting enzymes.Curr Opin Lipidol. 1998 Apr;9(2):99-102. doi: 10.1097/00041433-199804000-00004. Curr Opin Lipidol. 1998. PMID: 9559265 Review.
-
Structure and function of eukaryotic proprotein processing enzymes of the subtilisin family of serine proteases.Crit Rev Oncog. 1993;4(2):115-36. Crit Rev Oncog. 1993. PMID: 8420571 Review.
Cited by
-
Regulating the regulator: post-translational modification of RAS.Nat Rev Mol Cell Biol. 2011 Dec 22;13(1):39-51. doi: 10.1038/nrm3255. Nat Rev Mol Cell Biol. 2011. PMID: 22189424 Free PMC article. Review.
-
Plasma membrane localization of Ras requires class C Vps proteins and functional mitochondria in Saccharomyces cerevisiae.Mol Cell Biol. 2006 Apr;26(8):3243-55. doi: 10.1128/MCB.26.8.3243-3255.2006. Mol Cell Biol. 2006. PMID: 16581797 Free PMC article.
-
Lamin A/C Cardiomyopathies: Current Understanding and Novel Treatment Strategies.Curr Treat Options Cardiovasc Med. 2017 Mar;19(3):21. doi: 10.1007/s11936-017-0520-z. Curr Treat Options Cardiovasc Med. 2017. PMID: 28299614 Review.
-
Membrane proteases in the bacterial protein secretion and quality control pathway.Microbiol Mol Biol Rev. 2012 Jun;76(2):311-30. doi: 10.1128/MMBR.05019-11. Microbiol Mol Biol Rev. 2012. PMID: 22688815 Free PMC article. Review.
-
Membrane targeting of Rab GTPases is influenced by the prenylation motif.Mol Biol Cell. 2003 May;14(5):1882-99. doi: 10.1091/mbc.e02-10-0639. Epub 2003 Feb 6. Mol Biol Cell. 2003. PMID: 12802062 Free PMC article.
References
-
- Clarke S. Annu Rev Biochem. 1992;61:355–386. - PubMed
-
- Zhang F L, Casey P J. Annu Rev Biochem. 1996;65:241–269. - PubMed
-
- McGrath J P, Varshavsky A. Nature (London) 1989;340:400–404. - PubMed
-
- Schafer W, Trueblood C, Yang C, Mayer M, Rosenberg S, Poulter C, Kim S-H, Rine J. Science. 1990;249:1133–1139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

