Nitric oxide dioxygenase: an enzymic function for flavohemoglobin
- PMID: 9724711
- PMCID: PMC27902
- DOI: 10.1073/pnas.95.18.10378
Nitric oxide dioxygenase: an enzymic function for flavohemoglobin
Abstract
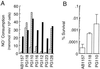
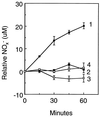
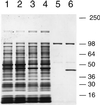
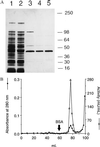
Nitric oxide (NO*) is a toxin, and various life forms appear to have evolved strategies for its detoxification. NO*-resistant mutants of Escherichia coli were isolated that rapidly consumed NO*. An NO*-converting activity was reconstituted in extracts that required NADPH, FAD, and O2, was cyanide-sensitive, and produced NO3-. This nitric oxide dioxygenase (NOD) contained 19 of 20 N-terminal amino acids identical to those of the E. coli flavohemoglobin. Furthermore, NOD activity was produced by the flavohemoglobin gene and was inducible by NO*. Flavohemoglobin/NOD-deficient mutants were also sensitive to growth inhibition by gaseous NO*. The results identify a function for the evolutionarily conserved flavohemoglobins and, moreover, suggest that NO* detoxification may be a more ancient function for the widely distributed hemoglobins, and associated methemoglobin reductases, than dioxygen transport and storage.
Figures

Similar articles
-
Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity.J Biol Chem. 2002 Mar 8;277(10):8166-71. doi: 10.1074/jbc.M110470200. Epub 2001 Dec 18. J Biol Chem. 2002. PMID: 11751864
-
Nitric oxide scavenging and detoxification by the Mycobacterium tuberculosis haemoglobin, HbN in Escherichia coli.Mol Microbiol. 2002 Sep;45(5):1303-14. doi: 10.1046/j.1365-2958.2002.03095.x. Mol Microbiol. 2002. PMID: 12207698
-
Type I flavohemoglobin of mycobacterium smegmatis is a functional nitric oxide dioxygenase.IUBMB Life. 2014 Jun;66(6):396-404. doi: 10.1002/iub.1275. Epub 2014 May 26. IUBMB Life. 2014. PMID: 24861678
-
Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases.J Inorg Biochem. 2005 Jan;99(1):247-66. doi: 10.1016/j.jinorgbio.2004.10.003. J Inorg Biochem. 2005. PMID: 15598505 Review.
-
Bacterial hemoglobins and flavohemoglobins: versatile proteins and their impact on microbiology and biotechnology.FEMS Microbiol Rev. 2003 Oct;27(4):525-45. doi: 10.1016/S0168-6445(03)00056-1. FEMS Microbiol Rev. 2003. PMID: 14550944 Review.
Cited by
-
Staphylococcus aureus SrrAB Affects Susceptibility to Hydrogen Peroxide and Co-Existence with Streptococcus sanguinis.PLoS One. 2016 Jul 21;11(7):e0159768. doi: 10.1371/journal.pone.0159768. eCollection 2016. PLoS One. 2016. PMID: 27441894 Free PMC article.
-
Nitric oxide and nitrous oxide turnover in natural and engineered microbial communities: biological pathways, chemical reactions, and novel technologies.Front Microbiol. 2012 Oct 23;3:372. doi: 10.3389/fmicb.2012.00372. eCollection 2012. Front Microbiol. 2012. PMID: 23109930 Free PMC article.
-
Fast ferrous heme-NO oxidation in nitric oxide synthases.FEBS J. 2009 Aug;276(16):4505-14. doi: 10.1111/j.1742-4658.2009.07157.x. FEBS J. 2009. PMID: 19691141 Free PMC article.
-
Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity.Plant Cell. 2004 Oct;16(10):2785-94. doi: 10.1105/tpc.104.025379. Epub 2004 Sep 14. Plant Cell. 2004. PMID: 15367716 Free PMC article.
-
Imidazole antibiotics inhibit the nitric oxide dioxygenase function of microbial flavohemoglobin.Antimicrob Agents Chemother. 2005 May;49(5):1837-43. doi: 10.1128/AAC.49.5.1837-1843.2005. Antimicrob Agents Chemother. 2005. PMID: 15855504 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

