A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit
- PMID: 9696837
- PMCID: PMC109968
- DOI: 10.1128/JVI.72.9.7407-7419.1998
A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit
Abstract
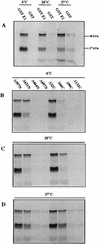
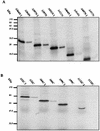
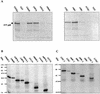
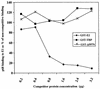
The human papillomavirus (HPV) E1 and E2 proteins bind cooperatively to the viral origin of replication (ori), forming an E1-E2-ori complex that is essential for initiation of DNA replication. All other replication proteins, including DNA polymerase alpha-primase (polalpha-primase), are derived from the host cell. We have carried out a detailed analysis of the interactions of HPV type 16 (HPV-16) E1 with E2, ori, and the four polalpha-primase subunits. Deletion analysis showed that a C-terminal region of E1 (amino acids [aa] 432 to 583 or 617) is required for E2 binding. HPV-16 E1 was unable to bind the ori in the absence of E2, but the same C-terminal domain of E1 was sufficient to tether E1 to the ori via E2. Of the polalpha-primase subunits, only p68 bound E1, and binding was competitive with E2. The E1 region required (aa 397 to 583) was the same as that required for E2 binding but additionally contained 34 N-terminal residues. In confirmation of these differences, we found that a monoclonal antibody, mapping adjacent to the N-terminal junction of the p68-binding region, blocked E1-p68 but not E1-E2 binding. Sequence alignments and secondary-structure prediction for HPV-16 E1 and other superfamily 3 (SF3) viral helicases closely parallel the mapping data in suggesting that aa 439 to 623 constitute a discrete helicase domain. Assuming a common nucleoside triphosphate-binding fold, we have generated a structural model of this domain based on the X-ray structures of the hepatitis C virus and Bacillus stearothermophilus (SF2) helicases. The modelling closely matches the deletion analysis in suggesting that this region of E1 is indeed a structural domain, and our results suggest that it is multifunctional and critical to several stages of HPV DNA replication.
Figures









Similar articles
-
Identification of domains of the HPV11 E1 protein required for DNA replication in vitro.Virology. 2000 Jun 20;272(1):137-50. doi: 10.1006/viro.2000.0328. Virology. 2000. PMID: 10873756
-
The carboxyl-terminal region of the human papillomavirus type 16 E1 protein determines E2 protein specificity during DNA replication.J Virol. 1998 Apr;72(4):3436-41. doi: 10.1128/JVI.72.4.3436-3441.1998. J Virol. 1998. PMID: 9525677 Free PMC article.
-
DNA replication specificity and functional E2 interaction of the E1 proteins of human papillomavirus types 1a and 18 are determined by their carboxyl-terminal halves.Virology. 1999 Apr 10;256(2):330-9. doi: 10.1006/viro.1999.9665. Virology. 1999. PMID: 10191198
-
The bacterial helicase-primase interaction: a common structural/functional module.Structure. 2005 Jun;13(6):839-44. doi: 10.1016/j.str.2005.04.006. Structure. 2005. PMID: 15939015 Free PMC article. Review.
-
The three-component helicase/primase complex of herpes simplex virus-1.Open Biol. 2021 Jun;11(6):210011. doi: 10.1098/rsob.210011. Epub 2021 Jun 9. Open Biol. 2021. PMID: 34102080 Free PMC article. Review.
Cited by
-
Papillomavirus E1 protein binds to and stimulates human topoisomerase I.J Virol. 2006 Feb;80(3):1584-7. doi: 10.1128/JVI.80.3.1584-1587.2006. J Virol. 2006. PMID: 16415033 Free PMC article.
-
Complete genome sequence of a novel extrachromosomal virus-like element identified in planarian Girardia tigrina.BMC Genomics. 2002 Jun 13;3:15. doi: 10.1186/1471-2164-3-15. Epub 2002 Jun 13. BMC Genomics. 2002. PMID: 12065025 Free PMC article.
-
Mitogen-activated protein kinases activate the nuclear localization sequence of human papillomavirus type 11 E1 DNA helicase to promote efficient nuclear import.J Virol. 2007 May;81(10):5066-78. doi: 10.1128/JVI.02480-06. Epub 2007 Mar 7. J Virol. 2007. PMID: 17344281 Free PMC article.
-
Characterization of the minimal DNA binding domain of the human papillomavirus e1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein.J Virol. 2003 May;77(9):5178-91. doi: 10.1128/jvi.77.9.5178-5191.2003. J Virol. 2003. PMID: 12692220 Free PMC article.
-
Human papillomavirus DNA replication compartments in a transient DNA replication system.J Virol. 1999 Feb;73(2):1001-9. doi: 10.1128/JVI.73.2.1001-1009.1999. J Virol. 1999. PMID: 9882301 Free PMC article.
References
-
- Bird L E, Subramanya H S, Wigley D B. Helicases: a unifying structural theme? Curr Opin Struct Biol. 1998;8:14–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

