The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms
- PMID: 9658103
- PMCID: PMC109835
- DOI: 10.1128/JVI.72.8.6581-6591.1998
The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms
Abstract
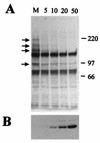
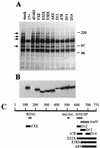
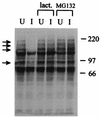
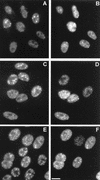
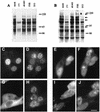
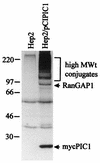
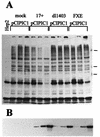
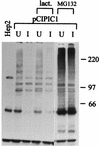
The small nuclear structures known as ND10 or PML nuclear bodies have been implicated in a variety of cellular processes including response to stress and interferons, oncogenesis, and viral infection, but little is known about their biochemical properties. Recently, a ubiquitin-specific protease enzyme (named HAUSP) and a ubiquitin-homology family protein (PIC1) have been found associated with ND10. HAUSP binds strongly to Vmw110, a herpesvirus regulatory protein which has the ability to disrupt ND10, while PIC1 was identified as a protein which interacts with PML, the prototype ND10 protein. We have investigated the role of ubiquitin-related pathways in the mechanism of ND10 disruption by Vmw110 and the effect of virus infection on PML stability. The results show that the disruption of ND10 during virus infection correlates with the loss of several PML isoforms and this process is dependent on active proteasomes. The PML isoforms that are most sensitive to virus infection correspond closely to those which have recently been identified as being covalently conjugated to PIC1. In addition, a large number of PIC1-protein conjugates can be detected following transfection of a PIC1 expression plasmid, and many of these are also eliminated in a Vmw110-dependent manner during virus infection. These observations provide a biochemical mechanism to explain the observed effects of Vmw110 on ND10 and suggest a simple yet powerful mechanism by which Vmw110 might function during virus infection.
Figures
Similar articles
-
Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins.J Virol. 2000 Nov;74(21):10006-17. doi: 10.1128/jvi.74.21.10006-10017.2000. J Virol. 2000. PMID: 11024129 Free PMC article.
-
A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein.EMBO J. 1997 Apr 1;16(7):1519-30. doi: 10.1093/emboj/16.7.1519. EMBO J. 1997. PMID: 9130697 Free PMC article.
-
A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein.EMBO J. 1997 Feb 3;16(3):566-77. doi: 10.1093/emboj/16.3.566. EMBO J. 1997. Corrected and republished in: EMBO J. 1997 Apr 1;16(7):1519-30. doi: 10.1093/emboj/16.7.1519 PMID: 9034339 Free PMC article. Corrected and republished.
-
PML and COP1--two proteins with much in common.Trends Biochem Sci. 2001 Jan;26(1):18-20. doi: 10.1016/s0968-0004(00)01732-1. Trends Biochem Sci. 2001. PMID: 11165511 Review.
-
The use of fluorescence microscopy to study the association between herpesviruses and intrinsic resistance factors.Viruses. 2011 Dec;3(12):2412-24. doi: 10.3390/v3122412. Epub 2011 Dec 7. Viruses. 2011. PMID: 22355446 Free PMC article. Review.
Cited by
-
Post-translational modifications of PML: consequences and implications.Front Oncol. 2013 Jan 4;2:210. doi: 10.3389/fonc.2012.00210. eCollection 2012. Front Oncol. 2013. PMID: 23316480 Free PMC article.
-
Single-cell analysis of Daxx and ATRX-dependent transcriptional repression.J Cell Sci. 2012 Nov 15;125(Pt 22):5489-501. doi: 10.1242/jcs.110148. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976303 Free PMC article.
-
Herpes simplex virus type 1 induces CD83 degradation in mature dendritic cells with immediate-early kinetics via the cellular proteasome.J Virol. 2007 Jun;81(12):6326-38. doi: 10.1128/JVI.02327-06. Epub 2007 Apr 11. J Virol. 2007. PMID: 17428858 Free PMC article.
-
Early expression of herpes simplex virus (HSV) proteins and reactivation of latent infection.Folia Microbiol (Praha). 2000;45(1):7-28. doi: 10.1007/BF02817445. Folia Microbiol (Praha). 2000. PMID: 11200675 Review.
-
PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication.J Cell Sci. 2011 Jan 15;124(Pt 2):280-91. doi: 10.1242/jcs.075390. Epub 2010 Dec 20. J Cell Sci. 2011. PMID: 21172801 Free PMC article.
References
-
- Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. - PubMed
-
- Boddy M N, Duprez E, Borden K L B, Freemont P S. Surface residue mutations of the PML RING finger domain alter the formation of nuclear-matrix-associated PML bodies. J Cell Sci. 1997;110:2197–2205. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

