Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-kappaB and Sp1 binding elements
- PMID: 9621017
- PMCID: PMC110216
- DOI: 10.1128/JVI.72.7.5589-5598.1998
Sequences just upstream of the simian immunodeficiency virus core enhancer allow efficient replication in the absence of NF-kappaB and Sp1 binding elements
Abstract
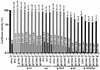
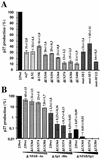
Large deletions of the upstream U3 sequences in the long terminal repeats (LTRs) of human immunodeficiency virus and simian immunodeficiency virus (SIV) accumulate in vivo in the absence of an intact nef gene. In the SIV U3 region, about 65 bp just upstream of the single NF-kappaB binding site always remained intact, and some evidence for a novel enhancer element in this region exists. We analyzed the transcriptional and replicative capacities of SIVmac239 mutants containing deletions or mutations in these upstream U3 sequences and/or the NF-kappaB and Sp1 binding sites. Even in the absence of 400 bp of upstream U3 sequences, the NF-kappaB site and all four Sp1 binding sites, the SIV promoter maintained about 15% of the wild-type LTR activity and was fully responsive to Tat activation in transient reporter assays. The effects of these deletions on virus production after transfection of COS-1 cells with full-length proviral constructs were much greater. Deletion of the upstream U3 sequences had no significant influence on viral replication when either the single NF-kappaB site or the Sp1 binding sites were intact. In contrast, the 26 bp of sequence located immediately upstream of the NF-kappaB site was essential for efficient replication when all core enhancer elements were deleted. A purine-rich site in this region binds specifically to the transcription factor Elf-1, a member of the ets proto-oncogene-encoded family. Our results indicate a high degree of functional redundancy in the SIVmac U3 region. Furthermore, we defined a novel regulatory element located immediately upstream of the NF-kappaB binding site that allows efficient viral replication in the absence of the entire core enhancer region.
Figures




Similar articles
-
Efficient transcription and replication of simian immunodeficiency virus in the absence of NF-kappaB and Sp1 binding elements.J Virol. 1996 May;70(5):3118-26. doi: 10.1128/JVI.70.5.3118-3126.1996. J Virol. 1996. PMID: 8627791 Free PMC article.
-
Induction of AIDS by simian immunodeficiency virus lacking NF-kappaB and SP1 binding elements.J Virol. 1997 Mar;71(3):1880-7. doi: 10.1128/JVI.71.3.1880-1887.1997. J Virol. 1997. PMID: 9032318 Free PMC article.
-
Upstream U3 sequences in simian immunodeficiency virus are selectively deleted in vivo in the absence of an intact nef gene.J Virol. 1994 Mar;68(3):2031-7. doi: 10.1128/JVI.68.3.2031-2037.1994. J Virol. 1994. PMID: 8107267 Free PMC article.
-
Human immunodeficiency virus type-1 transcription: role of the 5'-untranslated leader region (review).Int J Mol Med. 1998 May;1(5):875-81. doi: 10.3892/ijmm.1.5.875. Int J Mol Med. 1998. PMID: 9852310 Review.
-
Primate Lentiviruses Modulate NF-κB Activity by Multiple Mechanisms to Fine-Tune Viral and Cellular Gene Expression.Front Microbiol. 2017 Feb 14;8:198. doi: 10.3389/fmicb.2017.00198. eCollection 2017. Front Microbiol. 2017. PMID: 28261165 Free PMC article. Review.
Cited by
-
Simian immunodeficiency virus in which nef and U3 sequences do not overlap replicates efficiently in vitro and in vivo in rhesus macaques.J Virol. 2001 Sep;75(17):8137-46. doi: 10.1128/jvi.75.17.8137-8146.2001. J Virol. 2001. PMID: 11483759 Free PMC article.
-
NF-κB-Interacting Long Noncoding RNA Regulates HIV-1 Replication and Latency by Repressing NF-κB Signaling.J Virol. 2020 Aug 17;94(17):e01057-20. doi: 10.1128/JVI.01057-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581100 Free PMC article.
-
Disrupting surfaces of nef required for downregulation of CD4 and for enhancement of virion infectivity attenuates simian immunodeficiency virus replication in vivo.J Virol. 2000 Nov;74(21):9836-44. doi: 10.1128/jvi.74.21.9836-9844.2000. J Virol. 2000. PMID: 11024110 Free PMC article.
-
Simian and human immunodeficiency virus Nef proteins use different surfaces to downregulate class I major histocompatibility complex antigen expression.J Virol. 2000 Jun;74(12):5691-701. doi: 10.1128/jvi.74.12.5691-5701.2000. J Virol. 2000. PMID: 10823877 Free PMC article.
-
Development of minimal lentivirus vectors derived from simian immunodeficiency virus (SIVmac251) and their use for gene transfer into human dendritic cells.J Virol. 2000 Sep;74(18):8307-15. doi: 10.1128/jvi.74.18.8307-8315.2000. J Virol. 2000. PMID: 10954529 Free PMC article.
References
-
- Cullen B R. Use of eukaryotic expression technology in the functional analysis of cloned genes. Methods Enzymol. 1987;152:684–703. - PubMed
-
- Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Sonza S, Learmont J, Sullivan J S, Cunningham A, Dwyer D, Dowton D, Mills J. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. - PubMed
-
- Dewhurst S, Embretson J E, Anderson D C, Mullins J I, Fultz P N. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature. 1990;345:636–640. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

