Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria
- PMID: 9584158
- PMCID: PMC108899
- DOI: 10.1128/MCB.18.6.3173
Role of the negative charges in the cytosolic domain of TOM22 in the import of precursor proteins into mitochondria
Abstract
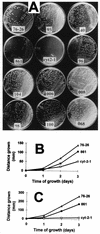
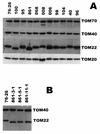
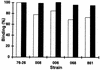
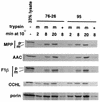
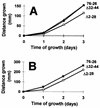
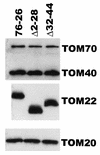
TOM22 is an essential mitochondrial outer membrane protein required for the import of precursor proteins into the organelles. The amino-terminal 84 amino acids of TOM22 extend into the cytosol and include 19 negatively and 6 positively charged residues. This region of the protein is thought to interact with positively charged presequences on mitochondrial preproteins, presumably via electrostatic interactions. We constructed a series of mutant derivatives of TOM22 in which 2 to 15 of the negatively charged residues in the cytosolic domain were changed to their corresponding amido forms. The mutant constructs were transformed into a sheltered Neurospora crassa heterokaryon bearing a tom22::hygromycin R disruption in one nucleus. All constructs restored viability to the disruption-carrying nucleus and gave rise to homokaryotic strains containing mutant tom22 alleles. Isolated mitochondria from three representative mutant strains, including the mutant carrying 15 neutralized residues (strain 861), imported precursor proteins at efficiencies comparable to those for wild-type organelles. Precursor binding studies with mitochondrial outer membrane vesicles from several of the mutant strains, including strain 861, revealed only slight differences from binding to wild-type vesicles. Deletion mutants lacking portions of the negatively charged region of TOM22 can also restore viability to the disruption-containing nucleus, but mutants lacking the entire region cannot. Taken together, these data suggest that an abundance of negative charges in the cytosolic domain of TOM22 is not essential for the binding or import of mitochondrial precursor proteins; however, other features in the domain are required.
Figures



Similar articles
-
Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria.Mol Cell Biol. 1996 Aug;16(8):4035-42. doi: 10.1128/MCB.16.8.4035. Mol Cell Biol. 1996. PMID: 8754801 Free PMC article.
-
The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences.Mol Cell Biol. 1997 Nov;17(11):6574-84. doi: 10.1128/MCB.17.11.6574. Mol Cell Biol. 1997. PMID: 9343421 Free PMC article.
-
An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22.J Biol Chem. 1998 May 8;273(19):11527-32. doi: 10.1074/jbc.273.19.11527. J Biol Chem. 1998. PMID: 9565567
-
Functions of outer membrane receptors in mitochondrial protein import.Biochim Biophys Acta. 2002 Sep 2;1592(1):3-14. doi: 10.1016/s0167-4889(02)00259-8. Biochim Biophys Acta. 2002. PMID: 12191763 Review.
-
The mitochondrial import machinery for preproteins.Crit Rev Biochem Mol Biol. 2001;36(3):291-336. doi: 10.1080/20014091074200. Crit Rev Biochem Mol Biol. 2001. PMID: 11450972 Review.
Cited by
-
Identification and functional analysis of human Tom22 for protein import into mitochondria.Mol Cell Biol. 2000 Oct;20(19):7205-13. doi: 10.1128/MCB.20.19.7205-7213.2000. Mol Cell Biol. 2000. PMID: 10982837 Free PMC article.
-
Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane.Proc Natl Acad Sci U S A. 1999 Mar 30;96(7):3634-9. doi: 10.1073/pnas.96.7.3634. Proc Natl Acad Sci U S A. 1999. PMID: 10097089 Free PMC article.
-
Interactions of amyloidogenic proteins with mitochondrial protein import machinery in aging-related neurodegenerative diseases.Front Physiol. 2023 Nov 2;14:1263420. doi: 10.3389/fphys.2023.1263420. eCollection 2023. Front Physiol. 2023. PMID: 38028797 Free PMC article. Review.
-
The loss in hydrophobic surface area resulting from a Leu to Val mutation at the N-terminus of the aldehyde dehydrogenase presequence prevents import of the protein into mitochondria.Protein Sci. 1999 Apr;8(4):890-6. doi: 10.1110/ps.8.4.890. Protein Sci. 1999. PMID: 10211835 Free PMC article.
-
Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex.J Cell Biol. 2001 Jan 22;152(2):289-300. doi: 10.1083/jcb.152.2.289. J Cell Biol. 2001. PMID: 11266446 Free PMC article.
References
-
- Austin B, Hall R M, Tyler B M. Optimized vectors for transformation of Neurospora crassa and Aspergillus nidulans to bleomycin and phleomycin resistance. Gene. 1990;93:157–162. - PubMed
-
- Ausubel R A, Brent R, Kingston R E, Moore D D, Seidman J G. Current protocols in molecular biology. New York, N.Y: Greene and Wiley Interscience; 1992.
-
- Bauer M F, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. - PubMed
-
- Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
