The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment
- PMID: 9560268
- PMCID: PMC20253
- DOI: 10.1073/pnas.95.9.5287
The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment
Abstract
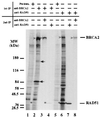
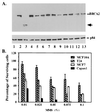
The BRCA2 gene was identified based on its involvement in familial breast cancer. The analysis of its sequence predicts that the gene encodes a protein with 3,418 amino acids but provides very few clues pointing to its biological function. In an attempt to address this question, specific antibodies were prepared that identified the gene product of BRCA2 as a 390-kDa nuclear protein. Furthermore, direct binding of human RAD51 to each of the four single 30-amino acid BRC repeats located at the 5' portion of exon 11 of BRCA2 was demonstrated. Such an interaction is significant, as BRCA2 and RAD51 can be reciprocally coimmunoprecipitated by each of the individual, specific antibodies and form complexes in vivo. Inferring from the function of RAD51 in DNA repair, human pancreatic cancer cells, Capan-1, expressing truncated BRCA2 were shown to be hypersensitive to methyl methanesulfonate (MMS) treatment. Exogenous expression of wild-type BRCA2, but not BRC-deleted mutants, in Capan-1 cells confers resistance to MMS treatment. These results suggest that the interaction between the BRC repeats of BRCA2 and RAD51 is critical for cellular response to DNA damage caused by MMS.
Figures
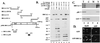


Similar articles
-
Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control.J Biol Chem. 1999 Nov 12;274(46):32931-5. doi: 10.1074/jbc.274.46.32931. J Biol Chem. 1999. PMID: 10551859
-
BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo.Cancer Res. 1999 Aug 1;59(15):3547-51. Cancer Res. 1999. PMID: 10446958
-
Distinct binding of BRCA2 BRC repeats to RAD51 generates differential DNA damage sensitivity.Nucleic Acids Res. 2016 Jun 20;44(11):5256-70. doi: 10.1093/nar/gkw242. Epub 2016 Apr 15. Nucleic Acids Res. 2016. PMID: 27084934 Free PMC article.
-
BRCA2 and homologous recombination.Breast Cancer Res. 2001;3(5):294-8. doi: 10.1186/bcr310. Epub 2001 Jul 11. Breast Cancer Res. 2001. PMID: 11597317 Free PMC article. Review.
-
Single-molecule imaging brings Rad51 nucleoprotein filaments into focus.Trends Cell Biol. 2010 May;20(5):269-76. doi: 10.1016/j.tcb.2010.02.004. Epub 2010 Mar 17. Trends Cell Biol. 2010. PMID: 20299221 Free PMC article. Review.
Cited by
-
Cancer-causing BRCA2 missense mutations disrupt an intracellular protein assembly mechanism to disable genome maintenance.Nucleic Acids Res. 2021 Jun 4;49(10):5588-5604. doi: 10.1093/nar/gkab308. Nucleic Acids Res. 2021. PMID: 33978741 Free PMC article.
-
Selective tumor killing based on specific DNA-damage response deficiencies.Cancer Biol Ther. 2012 Mar;13(5):239-46. doi: 10.4161/cbt.18921. Epub 2012 Mar 1. Cancer Biol Ther. 2012. PMID: 22258411 Free PMC article. Review.
-
Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8644-9. doi: 10.1073/pnas.151253498. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447276 Free PMC article.
-
Divergent binding mode for a protozoan BRC repeat to RAD51.Biochem J. 2022 May 27;479(10):1031-1043. doi: 10.1042/BCJ20220141. Biochem J. 2022. PMID: 35502837 Free PMC article.
-
BRCA1, BRCA2 and their possible function in DNA damage response.Br J Cancer. 1999 Dec;81(7):1099-102. doi: 10.1038/sj.bjc.6690814. Br J Cancer. 1999. PMID: 10584867 Free PMC article. Review. No abstract available.
References
-
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G, et al. Nature (London) 1995;378:789–792. - PubMed
-
- Tavtigian S V, Simard J, Rommens J, Couch F, Shattuckeidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppalyonnet D, et al. Nat Genet. 1996;12:333–337. - PubMed
-
- Wooster R, Neuhausen S L, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Science. 1994;265:2088–2090. - PubMed
-
- Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson J G, Tavtigian S V, Tulinius H, Ogmundsdottir H M, Eyfjörd J E. Nat Genet. 1996;13:117–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

