Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases
- PMID: 9560266
- PMCID: PMC20251
- DOI: 10.1073/pnas.95.9.5275
Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases
Abstract
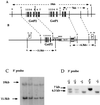
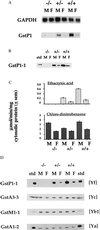
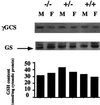
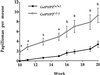
The activity of chemical carcinogens is a complex balance between metabolic activation by cytochrome P450 monooxygenases and detoxification by enzymes such as glutathione S-transferase (GST). Regulation of these proteins may have profound effects on carcinogenic activity, although it has proved impossible to ascribe the observed effects to the activity of a single protein. GstP appears to play a very important role in carcinogenesis, although the precise nature of its involvement is unclear. We have deleted the murine GstP gene cluster and established the effects on skin tumorigenesis induced by the polycyclic aromatic hydrocarbon 7, 12-dimethylbenz anthracene and the tumor promoting agent 12-O-tetradecanoylphorbol-13-acetate. After 20 weeks, a highly significant increase in the number of papillomas was found in the GstP1/P2 null mice [GstP1/P2(-/-) mice, 179 papillomas, mean 9.94 per animal vs. GstP1/P2(+/+) mice, 55 papillomas, mean 2.89 per animal, (P < 0.001)]. This difference in tumor incidence provides direct evidence that a single gene involved in drug metabolism can have a profound effect on tumorigenicity, and demonstrates that GstP may be an important determinant in cancer susceptibility, particularly in diseases where exposure to polycyclic aromatic hydrocarbons is involved, for instance in cigarette smoke-induced lung cancer.
Figures

Similar articles
-
Increased constitutive c-Jun N-terminal kinase signaling in mice lacking glutathione S-transferase Pi.J Biol Chem. 2003 Jun 20;278(25):22243-9. doi: 10.1074/jbc.M301211200. Epub 2003 Mar 19. J Biol Chem. 2003. PMID: 12646564
-
Increased resistance to acetaminophen hepatotoxicity in mice lacking glutathione S-transferase Pi.Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12741-5. doi: 10.1073/pnas.220176997. Proc Natl Acad Sci U S A. 2000. PMID: 11058152 Free PMC article.
-
Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma.Cancer Lett. 2005 Apr 28;221(2):135-43. doi: 10.1016/j.canlet.2004.08.028. Cancer Lett. 2005. PMID: 15808399
-
Pi-class glutathione S-transferase: regulation and function.Chem Biol Interact. 1998 Apr 24;111-112:69-82. doi: 10.1016/s0009-2797(97)00176-2. Chem Biol Interact. 1998. PMID: 9679544 Review.
-
The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance.Crit Rev Biochem Mol Biol. 1995;30(6):445-600. doi: 10.3109/10409239509083491. Crit Rev Biochem Mol Biol. 1995. PMID: 8770536 Review.
Cited by
-
Attenuation of lung fibrosis in mice with a clinically relevant inhibitor of glutathione-S-transferase π.JCI Insight. 2016 Jun 2;1(8):e85717. doi: 10.1172/jci.insight.85717. JCI Insight. 2016. PMID: 27358914 Free PMC article.
-
Glutathione S-transferase P protects against cyclophosphamide-induced cardiotoxicity in mice.Toxicol Appl Pharmacol. 2015 Jun 1;285(2):136-48. doi: 10.1016/j.taap.2015.03.029. Epub 2015 Apr 10. Toxicol Appl Pharmacol. 2015. PMID: 25868843 Free PMC article.
-
Nrf2: friend or foe for chemoprevention?Carcinogenesis. 2010 Jan;31(1):90-9. doi: 10.1093/carcin/bgp231. Epub 2009 Sep 30. Carcinogenesis. 2010. PMID: 19793802 Free PMC article. Review.
-
High-throughput library screening identifies two novel NQO1 inducers in human lung cells.Am J Respir Cell Mol Biol. 2012 Mar;46(3):365-71. doi: 10.1165/rcmb.2011-0301OC. Epub 2011 Oct 20. Am J Respir Cell Mol Biol. 2012. PMID: 22021338 Free PMC article.
-
Genetic or pharmacologic activation of Nrf2 signaling fails to protect against aflatoxin genotoxicity in hypersensitive GSTA3 knockout mice.Toxicol Sci. 2014 Jun;139(2):293-300. doi: 10.1093/toxsci/kfu056. Epub 2014 Mar 27. Toxicol Sci. 2014. PMID: 24675090 Free PMC article.
References
-
- Rushmore T H, Pickett C B. J Biol Chem. 1993;268:11475–11478. - PubMed
-
- Hayes J D, Strange R C. Free Radical Res. 1995;22:193–207. - PubMed
-
- Hayes J D, Pulford D J. Crit Rev Biochem Mol Biol. 1995;30:445–600. - PubMed
-
- Farber E. Cancer J Biochem Cell Biol. 1984;62:486–494. - PubMed
-
- Sato K, Kitahara A, Satoh K, Ishikawa T, Tatematsu M, Ito N. Gann. 1984;75:199–202. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

