Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate
- PMID: 9557745
- PMCID: PMC109685
- DOI: 10.1128/JVI.72.5.4478-4484.1998
Determinants of entry cofactor utilization and tropism in a dualtropic human immunodeficiency virus type 1 primary isolate
Abstract
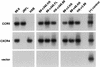
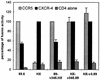
Human immunodeficiency virus type 1 strain 89.6 is a dualtropic isolate that replicates in macrophages and transformed T cells, and its envelope mediates CD4-dependent fusion and entry with CCR5, CXCR-4, and CCR3. To map determinants of cofactor utilization by 89.6 and determine the relationship between cofactor use and tropism, we analyzed recombinants generated between 89.6 and T-cell-tropic (HXB) or macrophage-tropic (JRFL) strains. These chimeras showed that regions of 89.6 env outside V3 through V5 determine CXCR-4 utilization and T-cell line tropism as well as CCR5 utilization and macrophage tropism. However, the 89.6 env V3 domain also conferred on HXB the ability to use CCR5 for fusion and entry but not the ability to establish productive macrophage infection. CCR3 use was conferred on HXB by 89.6 env V3 or V3 through V5 sequences. While replacement of the 89.6 V3 through V5 region with HXB sequences abrogated CCR3 utilization, replacement of V3 or V4 through V5 separately did not. Thus, CCR3 use is determined by sequences within V3 through V5 and most likely can be conferred by either the V3 or the V4 through V5 domains. These results indicate that cofactor utilization and tropism in this dualtropic isolate are determined by complex interactions among multiple env segments, that distinct regions of the Env glycoprotein may be important for utilization of different chemokine receptors, and that determinants in addition to cofactor usage participate in postentry stages in the virus replication cycle that contribute to target cell tropism.
Figures

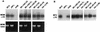

Similar articles
-
Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4.J Virol. 1998 Mar;72(3):2509-15. doi: 10.1128/JVI.72.3.2509-2515.1998. J Virol. 1998. PMID: 9499115 Free PMC article.
-
Role of V3 independent domains on a dualtropic human immunodeficiency virus type 1 (HIV-1) envelope gp120 in CCR5 coreceptor utilization and viral infectivity.Microbiol Immunol. 2001;45(7):521-30. doi: 10.1111/j.1348-0421.2001.tb02653.x. Microbiol Immunol. 2001. PMID: 11529558
-
V3-independent determinants of macrophage tropism in a primary human immunodeficiency virus type 1 isolate.J Virol. 1995 Mar;69(3):1755-61. doi: 10.1128/JVI.69.3.1755-1761.1995. J Virol. 1995. PMID: 7853514 Free PMC article.
-
HIV-1 envelope-receptor interactions required for macrophage infection and implications for current HIV-1 cure strategies.J Leukoc Biol. 2014 Jan;95(1):71-81. doi: 10.1189/jlb.0713368. Epub 2013 Oct 24. J Leukoc Biol. 2014. PMID: 24158961 Free PMC article. Review.
-
Asymmetric HIV-1 co-receptor use and replication in CD4(+) T lymphocytes.J Transl Med. 2011 Jan 27;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1479-5876-9-S1-S8. J Transl Med. 2011. PMID: 21284907 Free PMC article. Review.
Cited by
-
Envelope variants from women recently infected with clade A human immunodeficiency virus type 1 confer distinct phenotypes that are discerned by competition and neutralization experiments.J Virol. 2003 Aug;77(15):8448-61. doi: 10.1128/jvi.77.15.8448-8461.2003. J Virol. 2003. PMID: 12857914 Free PMC article.
-
Host range of small-ruminant lentivirus cytopathic variants determined with a selectable caprine arthritis- encephalitis virus pseudotype system.J Virol. 2001 Aug;75(16):7384-91. doi: 10.1128/JVI.75.16.7384-7391.2001. J Virol. 2001. PMID: 11462010 Free PMC article.
-
Conservation of human immunodeficiency virus type 1 gp120 inner-domain sequences in lentivirus and type A and B retrovirus envelope surface glycoproteins.J Virol. 2001 Feb;75(4):2014-8. doi: 10.1128/JVI.75.4.2014-2018.2001. J Virol. 2001. PMID: 11160703 Free PMC article.
-
Complex positive selection pressures drive the evolution of HIV-1 with different co-receptor tropisms.Sci China Life Sci. 2010 Oct;53(10):1204-14. doi: 10.1007/s11427-010-4066-5. Epub 2010 Oct 17. Sci China Life Sci. 2010. PMID: 20953943 Free PMC article.
-
HIV-1 gp120 chemokine receptor-mediated signaling in human macrophages.Immunol Res. 2003;27(2-3):261-76. doi: 10.1385/IR:27:2-3:261. Immunol Res. 2003. PMID: 12857973 Review.
References
-
- Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11:(Suppl. A):S3–S16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

