Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen
- PMID: 9528763
- PMCID: PMC121421
- DOI: 10.1128/MCB.18.4.1919
Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen
Abstract
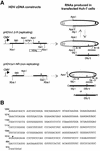
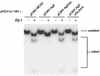
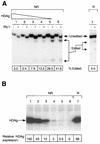
RNA editing at adenosine 1012 (amber/W site) in the antigenomic RNA of hepatitis delta virus (HDV) allows two essential forms of the viral protein, hepatitis delta antigen (HDAg), to be synthesized from a single open reading frame. Editing at the amber/W site is thought to be catalyzed by one of the cellular enzymes known as adenosine deaminases that act on RNA (ADARs). In vitro, the enzymes ADAR1 and ADAR2 deaminate adenosines within many different sequences of base-paired RNA. Since promiscuous deamination could compromise the viability of HDV, we wondered if additional deamination events occurred within the highly base paired HDV RNA. By sequencing cDNAs derived from HDV RNA from transfected Huh-7 cells, we determined that the RNA was not extensively modified at other adenosines. Approximately 0.16 to 0.32 adenosines were modified per antigenome during 6 to 13 days posttransfection. Interestingly, all observed non-amber/W adenosine modifications, which occurred mostly at positions that are highly conserved among naturally occurring HDV isolates, were found in RNAs that were also modified at the amber/W site. Such coordinate modification likely limits potential deleterious effects of promiscuous editing. Neither viral replication nor HDAg was required for the highly specific editing observed in cells. However, HDAg was found to suppress editing at the amber/W site when expressed at levels similar to those found during HDV replication. These data suggest HDAg may regulate amber/W site editing during virus replication.
Figures
Similar articles
-
Increased RNA editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2.J Virol. 2002 Apr;76(8):3819-27. doi: 10.1128/jvi.76.8.3819-3827.2002. J Virol. 2002. PMID: 11907222 Free PMC article.
-
RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure.J Virol. 2002 Aug;76(15):7385-97. doi: 10.1128/jvi.76.15.7385-7397.2002. J Virol. 2002. PMID: 12097551 Free PMC article.
-
Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2.J Virol. 2001 Sep;75(18):8547-55. doi: 10.1128/jvi.75.18.8547-8555.2001. J Virol. 2001. PMID: 11507200 Free PMC article.
-
RNA editing in hepatitis delta virus.Curr Top Microbiol Immunol. 2006;307:67-89. doi: 10.1007/3-540-29802-9_4. Curr Top Microbiol Immunol. 2006. PMID: 16903221 Review.
-
Control of ADAR1 editing of hepatitis delta virus RNAs.Curr Top Microbiol Immunol. 2012;353:123-43. doi: 10.1007/82_2011_146. Curr Top Microbiol Immunol. 2012. PMID: 21732238 Free PMC article. Review.
Cited by
-
Arginine-rich motifs are not required for hepatitis delta virus RNA binding activity of the hepatitis delta antigen.J Virol. 2013 Aug;87(15):8665-74. doi: 10.1128/JVI.00929-13. Epub 2013 Jun 5. J Virol. 2013. PMID: 23740973 Free PMC article.
-
Cellular Nuclear Export Factors TAP and Aly Are Required for HDAg-L-mediated Assembly of Hepatitis Delta Virus.J Biol Chem. 2016 Dec 9;291(50):26226-26238. doi: 10.1074/jbc.M116.754853. Epub 2016 Nov 2. J Biol Chem. 2016. PMID: 27807029 Free PMC article.
-
Epitranscriptomic marks: Emerging modulators of RNA virus gene expression.Wiley Interdiscip Rev RNA. 2020 May;11(3):e1576. doi: 10.1002/wrna.1576. Epub 2019 Nov 6. Wiley Interdiscip Rev RNA. 2020. PMID: 31694072 Free PMC article. Review.
-
Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes.Microbiol Mol Biol Rev. 1998 Dec;62(4):1415-34. doi: 10.1128/MMBR.62.4.1415-1434.1998. Microbiol Mol Biol Rev. 1998. PMID: 9841677 Free PMC article. Review.
-
Large hepatitis delta antigen is a novel clathrin adaptor-like protein.J Virol. 2007 Jun;81(11):5985-94. doi: 10.1128/JVI.02809-06. Epub 2007 Mar 21. J Virol. 2007. PMID: 17376909 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
