Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection
- PMID: 9525597
- PMCID: PMC109722
- DOI: 10.1128/JVI.72.4.2777-2787.1998
Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection
Abstract
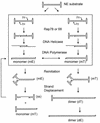
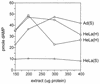
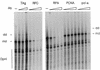
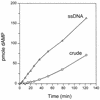
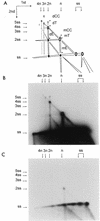
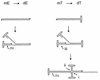
We previously reported the development of an in vitro adeno-associated virus (AAV) DNA replication system. The system required one of the p5 Rep proteins encoded by AAV (either Rep78 or Rep68) and a crude adenovirus (Ad)-infected HeLa cell cytoplasmic extract to catalyze origin of replication-dependent AAV DNA replication. However, in addition to fully permissive DNA replication, which occurs in the presence of Ad, AAV is also capable of partially permissive DNA replication in the absence of the helper virus in cells that have been treated with genotoxic agents. Limited DNA replication also occurs in the absence of Ad during the process of establishing a latent infection. In an attempt to isolate uninfected extracts that would support AAV DNA replication, we discovered that HeLa cell extracts grown to high density can occasionally display as much in vitro replication activity as Ad-infected extracts. This finding confirmed previous genetic analyses which suggested that no Ad-encoded proteins were absolutely essential for AAV DNA replication and that the uninfected extracts should be useful for studying the differences between helper-dependent and helper-independent AAV DNA replication. Using specific chemical inhibitors and monoclonal antibodies, as well as the fractionation of uninfected HeLa extracts, we identified several of the cellular enzymes involved in AAV DNA replication. They were the single-stranded DNA binding protein, replication protein A (RFA), the 3' primer binding complex, replication factor C (RFC), and proliferating cell nuclear antigen (PCNA). Consistent with the current model for AAV DNA replication, which requires only leading-strand DNA synthesis, we found no requirement for DNA polymerase alpha-primase. AAV DNA replication could be reconstituted with purified Rep78, RPA, RFC, and PCNA and a phosphocellulose chromatography fraction (IIA) that contained DNA polymerase activity. As both RFC and PCNA are known to be accessory proteins for polymerase delta and epsilon, we attempted to reconstitute AAV DNA replication by substituting either purified polymerase delta or polymerase epsilon for fraction IIA. These attempts were unsuccessful and suggested that some novel cellular protein or modification was required for AAV DNA replication that had not been previously identified. Finally, we also further characterized the in vitro DNA replication assay and demonstrated by two-dimensional (2D) gel electrophoresis that all of the intermediates commonly seen in vivo are generated in the in vitro system. The only difference was an accumulation of single-stranded DNA in vivo that was not seen in vitro. The 2D data also suggested that although both Rep78 and Rep68 can generate dimeric intermediates in vitro, Rep68 is more efficient in processing dimers to monomer duplex DNA. Regardless of the Rep that was used in vitro, we found evidence of an interaction between the elongation complex and the terminal repeats. Nicking at the terminal repeats of a replicating molecule appeared to be inhibited until after elongation was complete.
Figures



Similar articles
-
Purification of host cell enzymes involved in adeno-associated virus DNA replication.J Virol. 2007 Jun;81(11):5777-87. doi: 10.1128/JVI.02651-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360744 Free PMC article.
-
In vitro replication of adeno-associated virus DNA.J Virol. 1994 Feb;68(2):1128-38. doi: 10.1128/JVI.68.2.1128-1138.1994. J Virol. 1994. PMID: 8289342 Free PMC article.
-
High-level expression of adeno-associated virus (AAV) Rep78 or Rep68 protein is sufficient for infectious-particle formation by a rep-negative AAV mutant.J Virol. 1995 Nov;69(11):6880-5. doi: 10.1128/JVI.69.11.6880-6885.1995. J Virol. 1995. PMID: 7474103 Free PMC article.
-
The Interplay between Adeno-Associated Virus and its Helper Viruses.Viruses. 2020 Jun 19;12(6):662. doi: 10.3390/v12060662. Viruses. 2020. PMID: 32575422 Free PMC article. Review.
-
The adenovirus DNA-binding protein DBP.J Virol. 2024 Feb 20;98(2):e0188523. doi: 10.1128/jvi.01885-23. Epub 2024 Jan 10. J Virol. 2024. PMID: 38197632 Free PMC article. Review.
Cited by
-
Purification of host cell enzymes involved in adeno-associated virus DNA replication.J Virol. 2007 Jun;81(11):5777-87. doi: 10.1128/JVI.02651-06. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360744 Free PMC article.
-
Adeno-associated virus: from defective virus to effective vector.Virol J. 2005 May 6;2:43. doi: 10.1186/1743-422X-2-43. Virol J. 2005. PMID: 15877812 Free PMC article. Review.
-
Herpes simplex virus type 1/adeno-associated virus hybrid vectors.Open Virol J. 2010 Jun 18;4:109-22. doi: 10.2174/1874357901004030109. Open Virol J. 2010. PMID: 20811580 Free PMC article.
-
Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location.Viruses. 2020 Jan 29;12(2):151. doi: 10.3390/v12020151. Viruses. 2020. PMID: 32013091 Free PMC article. Review.
-
Using viral vectors as gene transfer tools (Cell Biology and Toxicology Special Issue: ETCS-UK 1 day meeting on genetic manipulation of cells).Cell Biol Toxicol. 2010 Feb;26(1):1-20. doi: 10.1007/s10565-009-9139-5. Epub 2009 Oct 15. Cell Biol Toxicol. 2010. PMID: 19830583 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

