Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1
- PMID: 9525573
- PMCID: PMC109687
- DOI: 10.1128/JVI.72.4.2567-2576.1998
Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1
Abstract
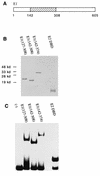
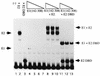
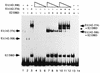
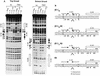
The bovine papillomavirus replication initiator protein E1 is an origin of replication (ori)-binding protein absolutely required for viral DNA replication. In the presence of the viral transcription factor E2, E1 binds to the ori and initiates DNA replication. To understand how the E1 initiator recognizes the ori and how E2 assists in this process, we have expressed and purified a 166-amino-acid fragment which corresponds to the minimal E1 DNA-binding domain (DBD). DNA binding studies using this protein demonstrate that the E1 DBD can bind to the palindromic E1 binding site in several forms but that binding of two monomers, each recognizing one half-site of the E1 palindrome, is the predominant form. This is reminiscent of the binding of the T-antigen DBD to the SV40 ori, and interestingly, the arrangement of E1 binding sites shows striking similarities to the arrangement of T-antigen binding sites in the SV40 ori even though the recognition sequences are unrelated. The E1 DBD is capable of interacting cooperatively with E2; however, the E2 DBD and not the E2 activation domain mediates this interaction. Furthermore, the E2 DBD stimulates binding of two monomers of the E1 DBD to the ori by binding cooperatively with one E1 monomer. Finally, we show that our results concerning the DNA-binding properties of the E1 DBD can be extended to full-length E1.
Figures




Similar articles
-
Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus.J Virol. 1997 Apr;71(4):2887-96. doi: 10.1128/JVI.71.4.2887-2896.1997. J Virol. 1997. PMID: 9060646 Free PMC article.
-
Two patches of amino acids on the E2 DNA binding domain define the surface for interaction with E1.J Virol. 2000 Feb;74(3):1506-12. doi: 10.1128/jvi.74.3.1506-1512.2000. J Virol. 2000. PMID: 10627562 Free PMC article.
-
Bovine papillomavirus type 1 DNA replication: the transcriptional activator E2 acts in vitro as a specificity factor.J Virol. 1997 Sep;71(9):6805-15. doi: 10.1128/JVI.71.9.6805-6815.1997. J Virol. 1997. PMID: 9261405 Free PMC article.
-
Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control.J Virol. 1998 Mar;72(3):1931-40. doi: 10.1128/JVI.72.3.1931-1940.1998. J Virol. 1998. PMID: 9499046 Free PMC article.
-
Papillomavirus E1 proteins: form, function, and features.Virus Genes. 2002 Jun;24(3):275-90. doi: 10.1023/a:1015336817836. Virus Genes. 2002. PMID: 12086149 Review.
Cited by
-
The papillomavirus E2 proteins.Virology. 2013 Oct;445(1-2):57-79. doi: 10.1016/j.virol.2013.06.006. Epub 2013 Jul 10. Virology. 2013. PMID: 23849793 Free PMC article. Review.
-
Genomic characterization of a novel Epsilonpapillomavirus associated with pigmented papillomas in a red deer (Cervus elaphus).Virus Genes. 2016 Oct;52(5):633-9. doi: 10.1007/s11262-016-1340-z. Epub 2016 May 6. Virus Genes. 2016. PMID: 27154332
-
Surface mutagenesis of the bovine papillomavirus E1 DNA binding domain reveals residues required for multiple functions related to DNA replication.J Virol. 2006 Aug;80(15):7491-9. doi: 10.1128/JVI.00435-06. J Virol. 2006. PMID: 16840329 Free PMC article.
-
Cellular topoisomerase I modulates origin binding by bovine papillomavirus type 1 E1.J Virol. 2006 May;80(9):4363-71. doi: 10.1128/JVI.80.9.4363-4371.2006. J Virol. 2006. PMID: 16611895 Free PMC article.
-
Sequential and ordered assembly of E1 initiator complexes on the papillomavirus origin of DNA replication generates progressive structural changes related to melting.Mol Cell Biol. 2002 Nov;22(21):7712-20. doi: 10.1128/MCB.22.21.7712-7720.2002. Mol Cell Biol. 2002. PMID: 12370317 Free PMC article.
References
-
- Chen, G., and A. Stenlund. Unpublished results.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

