Homeobox genes in the ribbonworm Lineus sanguineus: evolutionary implications
- PMID: 9501210
- PMCID: PMC19689
- DOI: 10.1073/pnas.95.6.3030
Homeobox genes in the ribbonworm Lineus sanguineus: evolutionary implications
Abstract
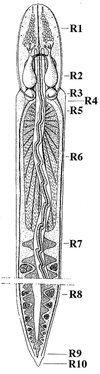
From our current understanding of the genetic basis of development and pattern formation in Drosophila and vertebrates it is commonly thought that clusters of Hox genes sculpt the morphology of animals in specific body regions. Based on Hox gene conservation throughout the animal kingdom it is proposed that these genes and their role in pattern formation evolved early during the evolution of metazoans. Knowledge of the history of Hox genes will lead to a better understanding of the role of Hox genes in the evolution of animal body plans. To infer Hox gene evolution, reliable data on lower chordates and invertebrates are crucial. Among the lower triploblasts, the body plan of the ribbonworm Lineus (nemertini) appears to be close to the common ancestral condition of protostomes and deuterostomes. In this paper we present the isolation and identification of Hox genes in Lineus sanguineus. We find that the Lineus genome contains a single cluster of at least six Hox genes: two anterior-class genes, three middle-class genes, and one posterior-class gene. Each of the genes can be definitely assigned to an ortholog group on the basis of its homeobox and its flanking sequences. The most closely related homeodomain sequences are invariably found among the mouse or Amphioxus orthologs, rather than Drosophila and other invertebrates. This suggests that the ribbonworms have diverged relatively little from the last common ancestors of protostomes and deuterostomes, the urbilateria.
Figures


Similar articles
-
Evolution of Antp-class genes and differential expression of Hydra Hox/paraHox genes in anterior patterning.Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4493-8. doi: 10.1073/pnas.97.9.4493. Proc Natl Acad Sci U S A. 2000. PMID: 10781050 Free PMC article.
-
Sipunculan ParaHox genes.Evol Dev. 2001 Jul-Aug;3(4):263-70. doi: 10.1046/j.1525-142x.2001.003004263.x. Evol Dev. 2001. PMID: 11478523
-
Evolution of invertebrate deuterostomes and Hox/ParaHox genes.Genomics Proteomics Bioinformatics. 2011 Jun;9(3):77-96. doi: 10.1016/S1672-0229(11)60011-9. Genomics Proteomics Bioinformatics. 2011. PMID: 21802045 Free PMC article. Review.
-
Coral comparative genomics reveal expanded Hox cluster in the cnidarian-bilaterian ancestor.Integr Comp Biol. 2012 Dec;52(6):835-41. doi: 10.1093/icb/ics098. Epub 2012 Jul 4. Integr Comp Biol. 2012. PMID: 22767488 Free PMC article.
-
Homeoboxes in sea anemones and other nonbilaterian animals: implications for the evolution of the Hox cluster and the zootype.Curr Top Dev Biol. 1998;40:211-54. doi: 10.1016/s0070-2153(08)60368-3. Curr Top Dev Biol. 1998. PMID: 9673852 Review. No abstract available.
Cited by
-
Hox genes pattern the anterior-posterior axis of the juvenile but not the larva in a maximally indirect developing invertebrate, Micrura alaskensis (Nemertea).BMC Biol. 2015 Apr 11;13:23. doi: 10.1186/s12915-015-0133-5. BMC Biol. 2015. PMID: 25888821 Free PMC article.
-
Hox gene survey in the chaetognath Spadella cephaloptera: evolutionary implications.Dev Genes Evol. 2003 Apr;213(3):142-8. doi: 10.1007/s00427-003-0306-z. Epub 2003 Mar 11. Dev Genes Evol. 2003. PMID: 12690453
-
HOX genes in the sepiolid squid Euprymna scolopes: implications for the evolution of complex body plans.Proc Natl Acad Sci U S A. 2002 Feb 19;99(4):2088-93. doi: 10.1073/pnas.042683899. Epub 2002 Feb 12. Proc Natl Acad Sci U S A. 2002. PMID: 11842209 Free PMC article.
-
Hox gene expression in larval development of the polychaetes Nereis virens and Platynereis dumerilii (Annelida, Lophotrochozoa).Dev Genes Evol. 2007 Jan;217(1):39-54. doi: 10.1007/s00427-006-0119-y. Epub 2006 Dec 19. Dev Genes Evol. 2007. PMID: 17180685
-
Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis.PLoS One. 2007 Jan 24;2(1):e153. doi: 10.1371/journal.pone.0000153. PLoS One. 2007. PMID: 17252055 Free PMC article.
References
-
- Slack J M W, Holland P W H, Graham C F. Nature (London) 1993;361:490–492. - PubMed
-
- Gehring W J. Science. 1987;236:1245–1252. - PubMed
-
- McGinnis W, Krumlauf R. Cell. 1992;68:283–302. - PubMed
-
- Salser S J, Kenyon C. Trends Genet. 1994;10:159–164. - PubMed
-
- Shankland M. BioEssays. 1994;16:801–808. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

