A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA
- PMID: 9501183
- PMCID: PMC19662
- DOI: 10.1073/pnas.95.6.2874
A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA
Abstract
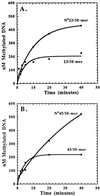
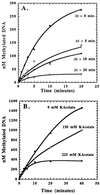
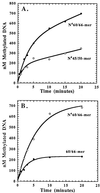
The kinetic properties of an adenine DNA methyltransferase involved in cell cycle regulation of Caulobacter crescentus have been elucidated by using defined unmethylated or hemimethylated DNA (DNAHM) substrates. Catalytic efficiency is significantly enhanced with a DNAHM substrate. Biphasic kinetic behavior during methyl incorporation is observed when unmethylated or DNAHM substrates are used, indicating that a step after chemistry limits enzyme turnover and is most likely the release of enzyme from methylated DNA product. The enzyme is thermally inactivated at 30 degrees C within 20 min; this process is substantially decreased in the presence of saturating concentrations of DNAHM, suggesting that the enzyme preferentially binds DNA before S-adenosylmethionine. The activity of the enzyme shows an unusual sensitivity to salt levels, apparently dissociating more rapidly from methylated DNA product as the salt level is decreased. The enzyme acts processively during methylation of specific DNA sequences, indicating a preferred order of product release in which S-adenosylhomocysteine is released from enzyme before fully methylated DNA. The kinetic behavior and activity of the enzyme are consistent with the temporal constraints during the cell cycle-regulated methylation of newly replicated chromosomal DNA.
Figures
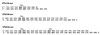


Similar articles
-
Caulobacter crescentus Cell Cycle-Regulated DNA Methyltransferase Uses a Novel Mechanism for Substrate Recognition.Biochemistry. 2017 Aug 1;56(30):3913-3922. doi: 10.1021/acs.biochem.7b00378. Epub 2017 Jul 18. Biochemistry. 2017. PMID: 28661661
-
The highly specific, cell cycle-regulated methyltransferase from Caulobacter crescentus relies on a novel DNA recognition mechanism.J Biol Chem. 2018 Dec 7;293(49):19038-19046. doi: 10.1074/jbc.RA118.005212. Epub 2018 Oct 15. J Biol Chem. 2018. PMID: 30323065 Free PMC article.
-
The Caulobacter crescentus DNA-(adenine-N6)-methyltransferase CcrM methylates DNA in a distributive manner.Nucleic Acids Res. 2012 Feb;40(4):1708-16. doi: 10.1093/nar/gkr768. Epub 2011 Sep 16. Nucleic Acids Res. 2012. PMID: 21926159 Free PMC article.
-
DNA methyltransferases and epigenetic regulation in bacteria.FEMS Microbiol Rev. 2016 Sep;40(5):575-91. doi: 10.1093/femsre/fuw023. Epub 2016 Jul 29. FEMS Microbiol Rev. 2016. PMID: 27476077 Review.
-
Epigenetic regulation of the bacterial cell cycle.Curr Opin Microbiol. 2009 Dec;12(6):722-9. doi: 10.1016/j.mib.2009.08.005. Epub 2009 Sep 23. Curr Opin Microbiol. 2009. PMID: 19783470 Review.
Cited by
-
N6-Methyladenine: A Conserved and Dynamic DNA Mark.Adv Exp Med Biol. 2016;945:213-246. doi: 10.1007/978-3-319-43624-1_10. Adv Exp Med Biol. 2016. PMID: 27826841 Free PMC article. Review.
-
Comparison of CcrM-dependent methylation in Caulobacter crescentus and Brucella abortus by nanopore sequencing.J Bacteriol. 2024 Jun 20;206(6):e0008324. doi: 10.1128/jb.00083-24. Epub 2024 May 9. J Bacteriol. 2024. PMID: 38722176 Free PMC article.
-
New antibiotics--resistance is futile.PLoS Biol. 2004 Feb;2(2):E53. doi: 10.1371/journal.pbio.0020053. Epub 2004 Feb 17. PLoS Biol. 2004. PMID: 14966545 Free PMC article.
-
Kinetic analysis of Yersinia pestis DNA adenine methyltransferase activity using a hemimethylated molecular break light oligonucleotide.PLoS One. 2007 Aug 29;2(8):e801. doi: 10.1371/journal.pone.0000801. PLoS One. 2007. PMID: 17726531 Free PMC article.
-
Getting in the loop: regulation of development in Caulobacter crescentus.Microbiol Mol Biol Rev. 2010 Mar;74(1):13-41. doi: 10.1128/MMBR.00040-09. Microbiol Mol Biol Rev. 2010. PMID: 20197497 Free PMC article. Review.
References
-
- Cheng X. Annu Rev Biophys Biomol Struct. 1995;24:293–318. - PubMed
-
- Braaten B, Nou X, Koltenbach L, Low D. Cell. 1994;76:577–588. - PubMed
-
- Gartler S M, Riggs A D. Annu Rev Genet. 1983;17:155–190. - PubMed
-
- Swain J L, Stewart T A, Leder P. Cell. 1987;50:719–727. - PubMed
-
- Li E, Bestor T, Jaenisch R. Cell. 1992;69:915–926. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

