Arginine methylation facilitates the nuclear export of hnRNP proteins
- PMID: 9499403
- PMCID: PMC316575
- DOI: 10.1101/gad.12.5.679
Arginine methylation facilitates the nuclear export of hnRNP proteins
Abstract
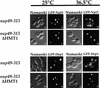
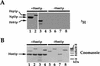
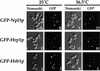
Eukaryotic mRNA processing and export is mediated by various heterogeneous nuclear ribonucleoproteins (hnRNPs). Many of these hnRNPs are methylated on arginine residues. In the yeast, Saccharomyces cerevisiae, the predominant enzyme responsible for arginine methylation is Hmt1p. Hmt1p methylates both Npl3p and Hrp1p, which are shuttling hnRNPs involved in mRNA processing and export. Here, we employ an in vivo nuclear export assay to show that arginine methylation is important for the nuclear export of these hnRNPs. Both Npl3p and Hrp1p fail to exit the nucleus in cells lacking Hmt1p, and overexpression of Hmt1p enhances Npl3p export. The export of a novel hnRNP-like protein, Hrb1p, which does not bind poly(A)+ RNA, however, is not affected by the lack of methylation. Furthermore, we find a genetic relationship between Hmt1p and cap-binding protein 80 (CBP80). Together, these findings establish that one biological role for arginine methylation is in facilitating the export of certain hnRNPs out of the nucleus.
Figures






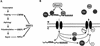
Similar articles
-
Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p.Mol Cell Biol. 2004 Dec;24(24):10742-56. doi: 10.1128/MCB.24.24.10742-10756.2004. Mol Cell Biol. 2004. PMID: 15572678 Free PMC article.
-
Novel RING finger proteins, Air1p and Air2p, interact with Hmt1p and inhibit the arginine methylation of Npl3p.J Biol Chem. 2000 Oct 20;275(42):32793-9. doi: 10.1074/jbc.M004560200. J Biol Chem. 2000. PMID: 10896665
-
Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p.J Biol Chem. 2002 Mar 8;277(10):7752-60. doi: 10.1074/jbc.M110053200. Epub 2002 Jan 4. J Biol Chem. 2002. PMID: 11779864
-
Nuclear export of mRNA.FEBS Lett. 2001 Jun 8;498(2-3):150-6. doi: 10.1016/s0014-5793(01)02482-6. FEBS Lett. 2001. PMID: 11412847 Review.
-
[Nuclear transport of hnRNP and mRNA].Tanpakushitsu Kakusan Koso. 2000 Oct;45(14):2378-87. Tanpakushitsu Kakusan Koso. 2000. PMID: 11051839 Review. Japanese. No abstract available.
Cited by
-
Yeast hnRNP-related proteins contribute to the maintenance of telomeres.Biochem Biophys Res Commun. 2012 Sep 14;426(1):12-7. doi: 10.1016/j.bbrc.2012.07.144. Epub 2012 Aug 10. Biochem Biophys Res Commun. 2012. PMID: 22902537 Free PMC article.
-
pUL69 of Human Cytomegalovirus Recruits the Cellular Protein Arginine Methyltransferase 6 via a Domain That Is Crucial for mRNA Export and Efficient Viral Replication.J Virol. 2015 Sep;89(18):9601-15. doi: 10.1128/JVI.01399-15. Epub 2015 Jul 15. J Virol. 2015. PMID: 26178996 Free PMC article.
-
Coupling pre-mRNA processing to transcription on the RNA factory assembly line.RNA Biol. 2013 Mar;10(3):380-90. doi: 10.4161/rna.23697. Epub 2013 Feb 7. RNA Biol. 2013. PMID: 23392244 Free PMC article. Review.
-
Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis.Biochemistry. 2007 Nov 20;46(46):13370-81. doi: 10.1021/bi701558t. Epub 2007 Oct 26. Biochemistry. 2007. PMID: 17960915 Free PMC article.
-
The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins.Mol Cell Biol. 2001 Dec;21(24):8289-300. doi: 10.1128/MCB.21.24.8289-8300.2001. Mol Cell Biol. 2001. PMID: 11713266 Free PMC article.
References
-
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. - PubMed
-
- Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG-NG-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74:969–976. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
