Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo
- PMID: 9499400
- PMCID: PMC316586
- DOI: 10.1101/gad.12.5.640
Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo
Abstract
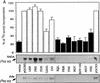
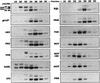
Several previously known transcription cofactors have been demonstrated in vitro recently to be histone acetyltransferases and deacetyltransferases, suggesting that remodeling of chromatin through histone acetylation plays a fundamental role in gene regulation. Clear evidence has not yet been obtained, however, to demonstrate that histone acetylation is required for gene activation in vivo. In this study we performed an alanine-scan mutagenesis through the HAT (histone acetyltransferase) domain identified previously by deletion mapping in recombinant yeast Gcn5. We identified multiple substitution mutations that eliminated completely Gcn5's ability to potentiate transcriptional activation in vivo. Strikingly, each of these mutations was also critical for free and nucleosomal histone acetylation by Gcn5 functioning within the native yeast HAT complexes, Ada, and SAGA. Moreover, the growth phenotypes of these mutations as measured by colony size and liquid growth assay closely tracked transcription and HAT activities. In contrast, mutations that did not affect in vivo function of Gcn5 were able to acetylate histones. These data argue strongly that acetylation is required for gene regulation by Gcn5 in vivo, and support previous arguments that nucleosomal histones are among the physiological substrates of acetylation by Gcn5.
Figures









Similar articles
-
Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex.Genes Dev. 1997 Jul 1;11(13):1640-50. doi: 10.1101/gad.11.13.1640. Genes Dev. 1997. PMID: 9224714
-
Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo.EMBO J. 1997 Feb 3;16(3):555-65. doi: 10.1093/emboj/16.3.555. EMBO J. 1997. PMID: 9034338 Free PMC article.
-
Identification and analysis of yeast nucleosomal histone acetyltransferase complexes.Methods. 1998 Aug;15(4):315-21. doi: 10.1006/meth.1998.0635. Methods. 1998. PMID: 9740719
-
Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
-
A SAGA of histone acetylation and gene expression.Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available.
Cited by
-
Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):8931-6. doi: 10.1073/pnas.96.16.8931. Proc Natl Acad Sci U S A. 1999. PMID: 10430873 Free PMC article.
-
The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae.Mol Cell Biol. 1999 Oct;19(10):6621-31. doi: 10.1128/MCB.19.10.6621. Mol Cell Biol. 1999. PMID: 10490601 Free PMC article.
-
Structure/function analysis of the phosphatidylinositol-3-kinase domain of yeast tra1.Genetics. 2007 Sep;177(1):151-66. doi: 10.1534/genetics.107.074476. Epub 2007 Jul 29. Genetics. 2007. PMID: 17660562 Free PMC article.
-
Systematic analysis of a conserved region of the aminoglycoside 6'-N-acetyltransferase type Ib.Antimicrob Agents Chemother. 2001 Dec;45(12):3287-92. doi: 10.1128/AAC.45.12.3287-3292.2001. Antimicrob Agents Chemother. 2001. PMID: 11709299 Free PMC article.
-
Plasmodium falciparum histone acetyltransferase, a yeast GCN5 homologue involved in chromatin remodeling.Eukaryot Cell. 2004 Apr;3(2):264-76. doi: 10.1128/EC.3.2.264-276.2004. Eukaryot Cell. 2004. PMID: 15075257 Free PMC article.
References
-
- Alland L, Muhle R, Hou Jr H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Bannister A, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. - PubMed
-
- Barlev N, Candau R, Wang L, Darpino P, Silverman N, Berger S. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
-
- Berger SL, Cress WD, Cress A, Triezenberg SJ, Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: Evidence for transcriptional adaptors. Cell. 1990;61:1199–1208. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
