Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro
- PMID: 9499085
- PMCID: PMC109524
- DOI: 10.1128/JVI.72.3.2265-2271.1998
Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro
Abstract
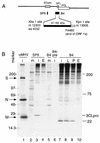
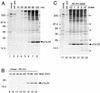
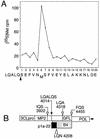
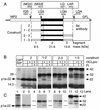
The 3C-like proteinase (3CLpro) of mouse hepatitis virus (MHV) is predicted to cleave at least 11 sites in the 803-kDa gene 1 polyprotein, resulting in maturation of proteinase, polymerase, and helicase proteins. However, most of these cleavage sites have not been experimentally confirmed and the proteins have not been identified in vitro or in virus-infected cells. We used specific antibodies to identify and characterize a 22-kDa protein (p1a-22) expressed from gene 1 in MHV A59-infected DBT cells. Processing of p1a-22 from the polyprotein began immediately after translation, but some processing continued for several hours. Amino-terminal sequencing of p1a-22 purified from MHV-infected cells showed that it was cleaved at a putative 3CLpro cleavage site, Gln_Ser4014 (where the underscore indicates the site of cleavage), that is located between the 3CLpro domain and the end of open reading frame (ORF) 1a. Subclones of this region of gene 1 were used to express polypeptides in vitro that contained one or more 3CLpro cleavage sites, and cleavage of these substrates by recombinant 3CLpro in vitro confirmed that amino-terminal cleavage of p1a-22 occurred at Gln_Ser4014. We demonstrated that the carboxy-terminal cleavage of the p1a-22 protein occurred at Gln_Asn4208, a sequence that had not been predicted as a site for cleavage by MHV 3CLpro. Our results demonstrate the usefulness of recombinant MHV 3CLpro in identifying and confirming cleavage sites within the gene 1 polyprotein. Based on our results, we predict that at least seven mature proteins are processed from the ORF 1a polyprotein by 3CLpro and suggest that additional noncanonical cleavage sites may be used by 3CLpro during processing of the gene 1 polyprotein.
Figures

Similar articles
-
Processing of the MHV-A59 gene 1 polyprotein by the 3C-like proteinase.Adv Exp Med Biol. 1998;440:121-7. doi: 10.1007/978-1-4615-5331-1_16. Adv Exp Med Biol. 1998. PMID: 9782273
-
Determinants of mouse hepatitis virus 3C-like proteinase activity.Virology. 1997 Apr 14;230(2):335-42. doi: 10.1006/viro.1997.8479. Virology. 1997. PMID: 9143289 Free PMC article.
-
Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59.Virology. 1996 Aug 15;222(2):375-82. doi: 10.1006/viro.1996.0434. Virology. 1996. PMID: 8806521 Free PMC article.
-
Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes.Virology. 1997 Apr 14;230(2):309-22. doi: 10.1006/viro.1997.8503. Virology. 1997. PMID: 9143287 Free PMC article.
-
Further requirements for cleavage by the murine coronavirus 3C-like proteinase: identification of a cleavage site within ORF1b.Virology. 1999 Oct 25;263(2):471-84. doi: 10.1006/viro.1999.9954. Virology. 1999. PMID: 10544119 Free PMC article.
Cited by
-
Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2.J Virol. 2003 Jul;77(13):7376-82. doi: 10.1128/jvi.77.13.7376-7382.2003. J Virol. 2003. PMID: 12805436 Free PMC article.
-
Dynamics of coronavirus replication-transcription complexes.J Virol. 2010 Feb;84(4):2134-49. doi: 10.1128/JVI.01716-09. Epub 2009 Dec 9. J Virol. 2010. PMID: 20007278 Free PMC article.
-
Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab.J Virol. 1999 Jan;73(1):177-85. doi: 10.1128/JVI.73.1.177-185.1999. J Virol. 1999. PMID: 9847320 Free PMC article.
-
Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication.J Virol. 2007 Nov;81(22):12323-36. doi: 10.1128/JVI.01506-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855519 Free PMC article.
-
Discovery of highly potent SARS-CoV-2 Mpro inhibitors based on benzoisothiazolone scaffold.Bioorg Med Chem Lett. 2022 Feb 15;58:128526. doi: 10.1016/j.bmcl.2022.128526. Epub 2022 Jan 5. Bioorg Med Chem Lett. 2022. PMID: 34998903 Free PMC article.
References
-
- Boursnell M F G, Brown T D K, Foulds I J, Green P F, Tomley F M, Binns M M. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. - PubMed
-
- Breedenbeek P J, Pachuk C J, Noten A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

