p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus
- PMID: 9499057
- PMCID: PMC109496
- DOI: 10.1128/JVI.72.3.2033-2039.1998
p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus
Abstract
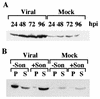
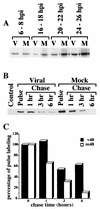
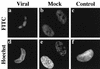
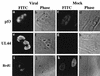
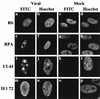
Previously, we reported that human cytomegalovirus (HCMV) infection of fibroblasts markedly affects p53 and other regulatory proteins and inhibits transit through the cell cycle (F. M. Jault, J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector, J. Virol. 69:6697-6704, 1995). Although the p53 steady-state levels are elevated throughout the infection, evidence suggests that the ability of p53 to transactivate some of its downstream targets is compromised. To elucidate the mechanisms governing the accumulation of p53, we examined the synthesis, stability, and localization of the protein in HCMV-infected fibroblasts. Synthesis of p53 was not increased in the infected cells during the first 24 h postinfection. In fact, pulse-chase experiments revealed that synthesis of p53 in infected fibroblasts was lower than in mock-infected cells. However, after an initial decay, the p53 was stabilized. In addition, beginning at approximately 30 h postinfection, p53 was localized to discrete foci within the nuclei of infected cells. The morphology of these foci suggested that they were replication centers. We confirmed that these are sites of DNA replication by demonstrating both incorporation of bromodeoxyuridine and localization of UL44 (the viral polymerase processivity factor) into these centers. The single-stranded DNA binding protein RPA was also sequestered. In contrast, Rb and HCMV IE1 72 remained distributed throughout the infected cell nuclei, indicating specific targeting of certain proteins. Taken together, our results provide two alternative mechanisms to account for the increased steady-state levels of p53 observed in HCMV-infected fibroblasts.
Figures
Similar articles
-
Dysregulation of cyclin E gene expression in human cytomegalovirus-infected cells requires viral early gene expression and is associated with changes in the Rb-related protein p130.J Virol. 2000 May;74(9):4192-206. doi: 10.1128/jvi.74.9.4192-4206.2000. J Virol. 2000. PMID: 10756032 Free PMC article.
-
The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10).J Virol. 1999 Dec;73(12):10458-71. doi: 10.1128/JVI.73.12.10458-10471.1999. J Virol. 1999. PMID: 10559364 Free PMC article.
-
Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection.J Virol. 2007 Feb;81(4):1934-50. doi: 10.1128/JVI.01670-06. Epub 2006 Dec 6. J Virol. 2007. PMID: 17151099 Free PMC article.
-
Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription.J Virol. 1998 May;72(5):3729-41. doi: 10.1128/JVI.72.5.3729-3741.1998. J Virol. 1998. PMID: 9557655 Free PMC article.
-
Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus.J Virol. 2003 Dec;77(24):13214-24. doi: 10.1128/jvi.77.24.13214-13224.2003. J Virol. 2003. PMID: 14645578 Free PMC article.
Cited by
-
Human cytomegalovirus pUL29/28 and pUL38 repression of p53-regulated p21CIP1 and caspase 1 promoters during infection.J Virol. 2013 Mar;87(5):2463-74. doi: 10.1128/JVI.01926-12. Epub 2012 Dec 12. J Virol. 2013. PMID: 23236067 Free PMC article.
-
The retinoblastoma tumor suppressor promotes efficient human cytomegalovirus lytic replication.J Virol. 2015 May;89(9):5012-21. doi: 10.1128/JVI.00175-15. Epub 2015 Feb 18. J Virol. 2015. PMID: 25694602 Free PMC article.
-
Human Cytomegalovirus nuclear egress and secondary envelopment are negatively affected in the absence of cellular p53.Virology. 2016 Oct;497:279-293. doi: 10.1016/j.virol.2016.07.021. Epub 2016 Aug 5. Virology. 2016. PMID: 27498410 Free PMC article.
-
Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis.J Virol. 2005 Oct;79(19):12205-17. doi: 10.1128/JVI.79.19.12205-12217.2005. J Virol. 2005. PMID: 16160147 Free PMC article.
-
Human cytomegalovirus IE1-72 protein interacts with p53 and inhibits p53-dependent transactivation by a mechanism different from that of IE2-86 protein.J Virol. 2009 Dec;83(23):12388-98. doi: 10.1128/JVI.00304-09. Epub 2009 Sep 23. J Virol. 2009. PMID: 19776115 Free PMC article.
References
-
- AbuBakar S, Au W W, Legator M S, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ Mol Mutagen. 1988;12:409–420. - PubMed
-
- Albrecht T, Fons M P, Bologh I, AbuBakar S, Deng C Z, Millinoff D. Metabolic and cellular effects of human cytomegalovirus infection. Transplant Proc. 1991;23:48–55. - PubMed
-
- Albrecht T, Fons M P, Deng C Z, Boldogh I. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology. 1997;230:48–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

