Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole
- PMID: 9490720
- PMCID: PMC2132703
- DOI: 10.1083/jcb.140.5.1063
Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole
Abstract
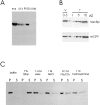
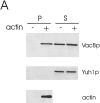
During each cell cycle, the yeast vacuole actively partitions between mother and daughter cells. This process requires actin, profilin, an unconventional myosin (Myo2p), and Vac8p. A mutant yeast strain, vac8, is defective in vacuole inheritance, specifically, in early vacuole migration. Vac8p is a 64-kD protein found on the vacuole membrane, a site consistent with its role in vacuole inheritance. Both myristoylation and palmitoylation are required for complete Vac8p localization. Interestingly, whereas myristoylation of Vac8p is not required for vacuole inheritance, palmitoylation is essential. Thus, palmitoylation appears to play a more direct role in vacuole inheritance. Most of the VAC8 sequence encodes 11 armadillo (Arm) repeats. Arm repeats are thought to mediate protein-protein interactions, and many Arm proteins have multiple functions. This is also true for Vac8p. In addition to its role in early vacuole inheritance, Vac8p is required to target aminopeptidase I from the cytoplasm to the vacuole. Mutant analysis demonstrates that Vac8p functions separately in these two processes. Vac8p cosediments with actin filaments. Vac8p is related to beta-catenin and plakoglobin, which connect a specific region of the plasma membrane to the actin cytoskeleton. In analogy, Vac8p may link the vacuole to actin during vacuole partitioning.
Figures









Similar articles
-
A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p.Mol Cell Biol. 2000 May;20(9):2984-95. doi: 10.1128/MCB.20.9.2984-2995.2000. Mol Cell Biol. 2000. PMID: 10757783 Free PMC article.
-
Yel013p (Vac8p), an armadillo repeat protein related to plakoglobin and importin alpha is associated with the yeast vacuole membrane.J Cell Sci. 1998 Oct;111 ( Pt 20):3109-18. doi: 10.1242/jcs.111.20.3109. J Cell Sci. 1998. PMID: 9739084
-
Palmitoylation plays a role in targeting Vac8p to specific membrane subdomains.Traffic. 2006 Oct;7(10):1378-87. doi: 10.1111/j.1600-0854.2006.00472.x. Traffic. 2006. PMID: 16978392
-
Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
-
Divide and multiply: organelle partitioning in yeast.Curr Opin Cell Biol. 2000 Aug;12(4):509-16. doi: 10.1016/s0955-0674(00)00124-1. Curr Opin Cell Biol. 2000. PMID: 10873824 Review.
Cited by
-
Autophagy in yeast: mechanistic insights and physiological function.Microbiol Mol Biol Rev. 2001 Sep;65(3):463-79, table of contents. doi: 10.1128/MMBR.65.3.463-479.2001. Microbiol Mol Biol Rev. 2001. PMID: 11528006 Free PMC article. Review.
-
Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p.Mol Biol Cell. 2000 Jul;11(7):2445-57. doi: 10.1091/mbc.11.7.2445. Mol Biol Cell. 2000. PMID: 10888680 Free PMC article.
-
The intricacy of nuclear membrane dynamics during nucleophagy.Nucleus. 2010 May-Jun;1(3):213-23. doi: 10.4161/nucl.1.3.11738. Epub 2010 Feb 16. Nucleus. 2010. PMID: 21327066 Free PMC article. Review.
-
Suppressors of mdm20 in yeast identify new alleles of ACT1 and TPM1 predicted to enhance actin-tropomyosin interactions.Genetics. 2000 Oct;156(2):523-34. doi: 10.1093/genetics/156.2.523. Genetics. 2000. PMID: 11014803 Free PMC article.
-
On the mechanism of protein palmitoylation.EMBO Rep. 2004 Nov;5(11):1053-7. doi: 10.1038/sj.embor.7400277. EMBO Rep. 2004. PMID: 15520806 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

