Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways
- PMID: 9482878
- PMCID: PMC19323
- DOI: 10.1073/pnas.95.5.2290
Human papillomavirus type 16 E6 and E7 oncogenes abrogate radiation-induced DNA damage responses in vivo through p53-dependent and p53-independent pathways
Abstract
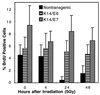
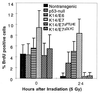
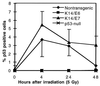
E6 and E7 oncoproteins from high risk human papillomaviruses (HPVs) transform cells in tissue culture and induce tumors in vivo. Both E6, which inhibits p53 functions, and E7, which inhibits pRb, can also abrogate growth arrest induced by DNA-damaging agents in cultured cells. In this study, we have used transgenic mice that express HPV-16 E6 or E7 in the epidermis to determine how these two proteins modulate DNA damage responses in vivo. Our results demonstrate that both E6 and E7 abrogate the inhibition of DNA synthesis in the epidermis after treatment with ionizing radiation. Increases in the levels of p53 and p21 proteins after irradiation were suppressed by E6 but not by E7. Through the study of p53-null mice, we found that radiation-induced growth arrest in the epidermis is mediated through both p53-dependent and p53-independent pathways. The abrogation of radiation responses in both E6 and E7 transgenic mice was more complete than was seen in the p53-null epidermis. We conclude that E6 and E7 each have the capacity to modulate p53-dependent as well as p53-independent cellular responses to radiation. Additionally, we found that the conserved region (CR) 1 and CR2 domains in E7 protein, which are involved in the inactivation of pRb function and required for E7's transforming function, were also required for E7 to modulate DNA damage responses in vivo. Thus pRb and/or pRb-like proteins likely mediate both p53-dependent and p53-independent responses to radiation.
Figures




Similar articles
-
Examination of the pRb-dependent and pRb-independent functions of E7 in vivo.J Virol. 2005 Sep;79(17):11392-402. doi: 10.1128/JVI.79.17.11392-11402.2005. J Virol. 2005. PMID: 16103190 Free PMC article.
-
Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways.Proc Natl Acad Sci U S A. 2000 Nov 7;97(23):12513-8. doi: 10.1073/pnas.97.23.12513. Proc Natl Acad Sci U S A. 2000. PMID: 11070078 Free PMC article.
-
p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein.Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5320-4. doi: 10.1073/pnas.91.12.5320. Proc Natl Acad Sci U S A. 1994. PMID: 8202487 Free PMC article.
-
Cellular targets of the oncoproteins encoded by the cancer associated human papillomaviruses.Princess Takamatsu Symp. 1991;22:239-48. Princess Takamatsu Symp. 1991. PMID: 1668886 Review.
-
Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products.Cancer Surv. 1992;12:197-217. Cancer Surv. 1992. PMID: 1322242 Review.
Cited by
-
Role of Rb-dependent and Rb-independent functions of papillomavirus E7 oncogene in head and neck cancer.Cancer Res. 2007 Dec 15;67(24):11585-93. doi: 10.1158/0008-5472.CAN-07-3007. Cancer Res. 2007. PMID: 18089787 Free PMC article.
-
Malignant Transformation of Fanconi Anemia Complementation Group D2-deficient (Fancd2 -/-) Hematopoietic Progenitor Cells by a Single HPV16 Oncogene.In Vivo. 2019 Mar-Apr;33(2):303-311. doi: 10.21873/invivo.11476. In Vivo. 2019. PMID: 30804107 Free PMC article.
-
Examination of the pRb-dependent and pRb-independent functions of E7 in vivo.J Virol. 2005 Sep;79(17):11392-402. doi: 10.1128/JVI.79.17.11392-11402.2005. J Virol. 2005. PMID: 16103190 Free PMC article.
-
Different responses of epidermal and hair follicular cells to radiation correlate with distinct patterns of p53 and p21 induction.Am J Pathol. 1999 Oct;155(4):1121-7. doi: 10.1016/S0002-9440(10)65215-7. Am J Pathol. 1999. PMID: 10514395 Free PMC article.
-
Validation of an HPV16-mediated carcinogenesis mouse model.In Vivo. 2014 Sep-Oct;28(5):761-7. In Vivo. 2014. PMID: 25189887 Free PMC article.
References
-
- zur Hausen H. Biochim Biophys Acta. 1996;1288:F55–F78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

