The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication
- PMID: 9445048
- PMCID: PMC124626
- DOI: 10.1128/JVI.72.2.1452-1461.1998
The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication
Abstract
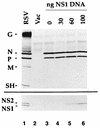
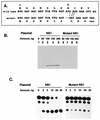
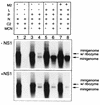
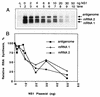
The NS1 protein (139 amino acids) is one of the two nonstructural proteins of human respiratory syncytial virus (RSV) and is encoded by a very abundant mRNA transcribed from the promoter-proximal RSV gene. The function of NS1 was unknown and was investigated here by using a reconstituted transcription and RNA replication system that involves a minireplicon and viral proteins (N, P, L and M2-1) expressed from separate cotransfected plasmids. Coexpression of the NS1 cDNA strongly inhibited transcription and RNA replication mediated by the RSV polymerase, even when the level of expressed NS1 protein was substantially below that observed in RSV-infected cells. The effect depended on synthesis of NS1 protein rather than NS1 RNA alone. Transcription and both steps of RNA replication, namely, synthesis of the antigenome and the genome, appeared to be equally sensitive to inhibition. The efficiency of encapsidation of the plasmid-derived minigenome was not altered by coexpression of NS1, indicating that the inhibition occurs at a later step. In two different dicistronic minigenomes, transcription of each gene was equally sensitive to inhibition by NS1. This suggested that the gradient of transcriptional polarity was unaffected and that the effect of NS1 instead probably involves an early event such as polymerase entry on the genome. NS1-mediated inhibition of transcription and RNA replication was not affected by coexpression of the M2 mRNA, which has two open reading frames encoding the transcriptional elongation factor M2-1 and the putative negative regulatory factor M2-2. The potent nature of the NS1-mediated inhibition suggests that negative regulation is an authentic function of the NS1 protein, albeit not necessarily the only one.
Figures




Similar articles
-
RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA.J Virol. 1995 Sep;69(9):5677-86. doi: 10.1128/JVI.69.9.5677-5686.1995. J Virol. 1995. PMID: 7637014 Free PMC article.
-
Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome.Virology. 1997 Sep 15;236(1):188-201. doi: 10.1006/viro.1997.8734. Virology. 1997. PMID: 9299631
-
Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription.J Virol. 1999 Jul;73(7):5852-64. doi: 10.1128/JVI.73.7.5852-5864.1999. J Virol. 1999. PMID: 10364337 Free PMC article.
-
An Unexpected Encounter: Respiratory Syncytial Virus Nonstructural Protein 1 Interacts with Mediator Subunit MED25.J Virol. 2022 Oct 12;96(19):e0129722. doi: 10.1128/jvi.01297-22. Epub 2022 Sep 14. J Virol. 2022. PMID: 36102648 Free PMC article. Review.
-
Structural Insights into the Respiratory Syncytial Virus RNA Synthesis Complexes.Viruses. 2021 May 5;13(5):834. doi: 10.3390/v13050834. Viruses. 2021. PMID: 34063087 Free PMC article. Review.
Cited by
-
Animal pneumoviruses: molecular genetics and pathogenesis.Clin Microbiol Rev. 2004 Apr;17(2):390-412. doi: 10.1128/CMR.17.2.390-412.2004. Clin Microbiol Rev. 2004. PMID: 15084507 Free PMC article. Review.
-
Contribution of Dendritic Cells in Protective Immunity against Respiratory Syncytial Virus Infection.Viruses. 2020 Jan 15;12(1):102. doi: 10.3390/v12010102. Viruses. 2020. PMID: 31952261 Free PMC article. Review.
-
Unity in diversity: shared mechanism of entry among paramyxoviruses.Prog Mol Biol Transl Sci. 2015;129:1-32. doi: 10.1016/bs.pmbts.2014.10.001. Epub 2014 Dec 1. Prog Mol Biol Transl Sci. 2015. PMID: 25595799 Free PMC article. Review.
-
The NS2 protein of human respiratory syncytial virus suppresses the cytotoxic T-cell response as a consequence of suppressing the type I interferon response.J Virol. 2006 Jun;80(12):5958-67. doi: 10.1128/JVI.00181-06. J Virol. 2006. PMID: 16731934 Free PMC article.
-
Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins.J Virol. 2003 Aug;77(15):8426-39. doi: 10.1128/jvi.77.15.8426-8439.2003. J Virol. 2003. PMID: 12857912 Free PMC article.
References
-
- Alansari H, Potgeiter L N D. Nucleotide and predicted amino acid sequence analysis of the ovine respiratory syncytial virus nonstructural 1C and 1B genes and the small hydrophobic protein gene. J Gen Virol. 1994;75:401–404. - PubMed
-
- Chomczynski P. One hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal Biochem. 1992;20:134–139. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

