A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis
- PMID: 9418873
- PMCID: PMC121488
- DOI: 10.1128/MCB.18.1.260
A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis
Abstract
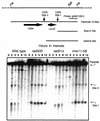
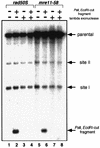
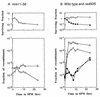
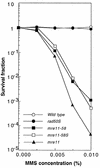
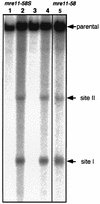
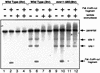
Using complementation tests and nucleotide sequencing, we showed that the rad58-4 mutation was an allele of the MRE11 gene and have renamed the mutation mre11-58. Two amino acid changes from the wild-type sequence were identified; one is located at a conserved site of a phosphodiesterase motif, and the other is a homologous amino acid change at a nonconserved site. Unlike mre11 null mutations, the mre11-58 mutation allowed meiosis-specific double-strand DNA breaks (DSBs) to form at recombination hot spots but failed to process those breaks. DSB ends of this mutant were resistant to lambda exonuclease treatment. These phenotypes are similar to those of rad50S mutants. In contrast to rad50S, however, mre11-58 was highly sensitive to methyl methanesulfonate treatment. DSB end processing induced by HO endonuclease was suppressed in both mre11-58 and the mre11 disruption mutant. We constructed a new mre11 mutant that contains only the phosphodiesterase motif mutation of the Mre11-58 protein and named it mre11-58S. This mutant showed the same phenotypes observed in mre11-58, suggesting that the phosphodiesterase consensus sequence is important for nucleolytic processing of DSB ends during both mitosis and meiosis.
Figures

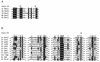

Similar articles
-
Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae.Genetics. 1995 Apr;139(4):1521-32. doi: 10.1093/genetics/139.4.1521. Genetics. 1995. PMID: 7789757 Free PMC article.
-
Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination.EMBO J. 1998 Nov 2;17(21):6412-25. doi: 10.1093/emboj/17.21.6412. EMBO J. 1998. PMID: 9799249 Free PMC article.
-
Functions of the yeast meiotic recombination genes, MRE11 and MRE2.Adv Biophys. 1995;31:67-76. doi: 10.1016/0065-227x(95)99383-z. Adv Biophys. 1995. PMID: 7625279 Review.
-
mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis.Genes Dev. 1997 Sep 1;11(17):2272-90. doi: 10.1101/gad.11.17.2272. Genes Dev. 1997. PMID: 9303542 Free PMC article.
-
The multiple roles of the Mre11 complex for meiotic recombination.Chromosome Res. 2007;15(5):551-63. doi: 10.1007/s10577-007-1147-9. Chromosome Res. 2007. PMID: 17674145 Review.
Cited by
-
Both conserved and non-conserved regions of Spo11 are essential for meiotic recombination initiation in yeast.Mol Genet Genomics. 2006 Oct;276(4):313-21. doi: 10.1007/s00438-006-0143-7. Epub 2006 Jul 1. Mol Genet Genomics. 2006. PMID: 16816949
-
Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage.Genes Dev. 2006 Sep 1;20(17):2437-49. doi: 10.1101/gad.1440206. Genes Dev. 2006. PMID: 16951256 Free PMC article.
-
Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks.Nat Struct Mol Biol. 2010 Dec;17(12):1478-85. doi: 10.1038/nsmb.1957. Epub 2010 Nov 21. Nat Struct Mol Biol. 2010. PMID: 21102445 Free PMC article.
-
Functional and structural insights into the MRX/MRN complex, a key player in recognition and repair of DNA double-strand breaks.Comput Struct Biotechnol J. 2020 May 16;18:1137-1152. doi: 10.1016/j.csbj.2020.05.013. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32489527 Free PMC article. Review.
-
The Msh5 complex shows homeostatic localization in response to DNA double-strand breaks in yeast meiosis.Front Cell Dev Biol. 2023 May 18;11:1170689. doi: 10.3389/fcell.2023.1170689. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37274743 Free PMC article.
References
-
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. - PubMed
-
- Baum B. Mre11 and S. pombe Rad32 are phosphoesterases. Yeast Update Newsl. 1995;3.7:1.
-
- Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. An atypical topoisomerase II from archaea with implications for meioric recombination. Nature. 1997;386:414–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
