The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX
- PMID: 9418860
- PMCID: PMC121463
- DOI: 10.1128/MCB.18.1.122
The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX
Abstract
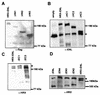
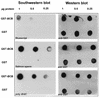
The HRX gene (also called MLL, ALL-1, and Htrx) at chromosome band 11q23 is associated with specific subsets of acute leukemias through translocations that result in its fusion with a variety of heterologous partners. Two of these partners, ENL and AF9, code for proteins that are highly similar to each other and as fusions with HRX induce myeloid leukemias in mice as demonstrated by retroviral gene transfer and knock-in experiments, respectively. In the present study, a structure-function analysis was performed to determine the molecular requirements for in vitro immortalization of murine myeloid cells by HRX-ENL. Deletions of either the AT hook motifs or the methyltransferase homology domain of HRX substantially impaired the transforming effects of HRX-ENL. The methyltransferase homology domain was shown to bind non-sequence specifically to DNA in vitro, providing evidence that the full transforming activity of HRX-ENL requires multiple DNA binding structures in HRX. The carboxy-terminal 84 amino acids of ENL, which encode two predicted helical structures highly conserved in AF9, were necessary and sufficient for transformation when they were fused to HRX. Similarly, mutations that deleted one or both of these conserved helices completely abrogated the transcriptional activation properties of ENL. This finding correlates, for the first time, a biological function of an HRX fusion partner with the transforming activity of the chimeric proteins. Our studies support a model in which HRX-ENL induces myeloid transformation by deregulating subordinate genes through a gain of function contributed by the transcriptional effector properties of ENL.
Figures



Similar articles
-
ENL, the gene fused with HRX in t(11;19) leukemias, encodes a nuclear protein with transcriptional activation potential in lymphoid and myeloid cells.Blood. 1994 Sep 15;84(6):1747-52. Blood. 1994. PMID: 8080983
-
Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL.EMBO J. 1997 Jul 16;16(14):4226-37. doi: 10.1093/emboj/16.14.4226. EMBO J. 1997. PMID: 9250666 Free PMC article.
-
The HRX proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures.Blood. 1997 May 1;89(9):3361-70. Blood. 1997. PMID: 9129043
-
Rearrangements involving chromosome band 11Q23 in acute leukaemia.Semin Cancer Biol. 1993 Dec;4(6):377-85. Semin Cancer Biol. 1993. PMID: 8142623 Review.
-
The structure of the human ALL-1/MLL/HRX gene.Leuk Lymphoma. 1997 Nov;27(5-6):417-28. doi: 10.3109/10428199709058308. Leuk Lymphoma. 1997. PMID: 9477123 Review.
Cited by
-
Targeting genetic alterations in protein methyltransferases for personalized cancer therapeutics.Oncogene. 2013 Feb 21;32(8):939-46. doi: 10.1038/onc.2012.552. Epub 2012 Nov 19. Oncogene. 2013. PMID: 23160372 Free PMC article. Review.
-
The molecular biology of mixed lineage leukemia.Haematologica. 2009 Jul;94(7):984-93. doi: 10.3324/haematol.2008.002436. Epub 2009 Jun 16. Haematologica. 2009. PMID: 19535349 Free PMC article. Review.
-
The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain.Mol Cell Biol. 1999 Jun;19(6):3931-9. doi: 10.1128/MCB.19.6.3931. Mol Cell Biol. 1999. PMID: 10330133 Free PMC article.
-
The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation.Nucleic Acids Res. 2002 Feb 15;30(4):958-65. doi: 10.1093/nar/30.4.958. Nucleic Acids Res. 2002. PMID: 11842107 Free PMC article.
-
Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9.Genes Dev. 2003 Sep 15;17(18):2298-307. doi: 10.1101/gad.1111603. Epub 2003 Sep 2. Genes Dev. 2003. PMID: 12952893 Free PMC article.
References
-
- Ashar H R, Fejzo M S, Tkachenk A, Zhou X, Fletcher J A, Weremowicz S, Morton C C, Chada K K. Disruption of the architectural factor HMGI-C: DNA-binding AT-hook motifs fused in lipomas to distinct transcriptional regulatory domains. Cell. 1995;82:57–65. - PubMed
-
- Bagga R, Emerson B M. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. - PubMed
-
- Bernard O A, Berger R. Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer. 1995;13:75–85. - PubMed
-
- Breen T R, Harte P J. trithorax regulates multiple homeotic genes in the bithorax and antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development. 1993;117:119–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
