The O-glycosylated stalk domain is required for apical sorting of neurotrophin receptors in polarized MDCK cells
- PMID: 9362511
- PMCID: PMC2139957
- DOI: 10.1083/jcb.139.4.929
The O-glycosylated stalk domain is required for apical sorting of neurotrophin receptors in polarized MDCK cells
Abstract
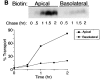
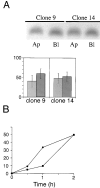
Delivery of newly synthesized membrane-spanning proteins to the apical plasma membrane domain of polarized MDCK epithelial cells is dependent on yet unidentified sorting signals present in the luminal domains of these proteins. In this report we show that structural information for apical sorting of transmembrane neurotrophin receptors (p75(NTR)) is localized to a juxtamembrane region of the extracellular domain that is rich in O-glycosylated serine/threonine residues. An internal deletion of 50 amino acids that removes this stalk domain from p75(NTR) causes the protein to be sorted exclusively of the basolateral plasma membrane. Basolateral sorting stalk-minus p75(NTR) does not occur by default, but requires sequences present in the cytoplasmic domain. The stalk domain is also required for apical secretion of a soluble form of p75(NTR), providing the first demonstration that the same domain can mediate apical sorting of both a membrane-anchored as well as secreted protein. However, the single N-glycan present on p75(NTR) is not required for apical sorting of either transmembrane or secreted forms.
Figures




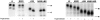




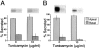


Similar articles
-
Role of the membrane-proximal O-glycosylation site in sorting of the human receptor for neurotrophins to the apical membrane of MDCK cells.Exp Cell Res. 2002 Feb 15;273(2):178-86. doi: 10.1006/excr.2001.5442. Exp Cell Res. 2002. PMID: 11822873
-
Putative O-glycosylation sites and a membrane anchor are necessary for apical delivery of the human neurotrophin receptor in Caco-2 cells.J Biol Chem. 1998 Nov 13;273(46):30263-70. doi: 10.1074/jbc.273.46.30263. J Biol Chem. 1998. PMID: 9804786
-
Multiple motifs regulate apical sorting of p75 via a mechanism that involves dimerization and higher-order oligomerization.Mol Biol Cell. 2013 Jun;24(12):1996-2007. doi: 10.1091/mbc.E13-02-0078. Epub 2013 May 1. Mol Biol Cell. 2013. PMID: 23637462 Free PMC article.
-
Structural Characterization of the p75 Neurotrophin Receptor: A Stranger in the TNFR Superfamily.Vitam Horm. 2017;104:57-87. doi: 10.1016/bs.vh.2016.10.007. Epub 2016 Nov 29. Vitam Horm. 2017. PMID: 28215307 Review.
-
Role of N- and O-glycans in polarized biosynthetic sorting.Am J Physiol Cell Physiol. 2006 Jan;290(1):C1-C10. doi: 10.1152/ajpcell.00333.2005. Am J Physiol Cell Physiol. 2006. PMID: 16338974 Review.
Cited by
-
O-glycosylation as a sorting determinant for cell surface delivery in yeast.Mol Biol Cell. 2004 Apr;15(4):1533-43. doi: 10.1091/mbc.e03-07-0511. Epub 2004 Jan 23. Mol Biol Cell. 2004. PMID: 14742720 Free PMC article.
-
Sodium pump localization in epithelia.J Bioenerg Biomembr. 2007 Dec;39(5-6):373-8. doi: 10.1007/s10863-007-9100-3. J Bioenerg Biomembr. 2007. PMID: 17972022 Review.
-
Secretory cargo composition affects polarized secretion in MDCK epithelial cells.Mol Cell Biochem. 2008 Mar;310(1-2):67-75. doi: 10.1007/s11010-007-9666-4. Epub 2007 Nov 30. Mol Cell Biochem. 2008. PMID: 18049865
-
Specific N-glycans direct apical delivery of transmembrane, but not soluble or glycosylphosphatidylinositol-anchored forms of endolyn in Madin-Darby canine kidney cells.Mol Biol Cell. 2004 Mar;15(3):1407-16. doi: 10.1091/mbc.e03-08-0550. Epub 2003 Dec 29. Mol Biol Cell. 2004. PMID: 14699065 Free PMC article.
-
Core-glycosylated mucin-like repeats from MUC1 are an apical targeting signal.J Biol Chem. 2011 Nov 11;286(45):39072-81. doi: 10.1074/jbc.M111.289504. Epub 2011 Sep 20. J Biol Chem. 2011. PMID: 21937430 Free PMC article.
References
-
- Baldwin AN, Shooter EM. Zone mapping of the binding domain of the rat low affinity nerve growth factor receptor by the introduction of novel N-glycosylation sites. J Biol Chem. 1995;270:4594–4602. - PubMed
-
- Baldwin AN, Bitler CM, Welcher AA, Shooter EM. Studies on the structure and binding properties of the cysteine-rich domain of rat low affinity nerve growth factor receptor (p75NGFR) J Biol Chem. 1992;267:8352–8359. - PubMed
-
- Blochberger TC, Sabatine JM, Lee YC, Hughey RP. O-linked glycosylation of rat renal g-glutamylpeptidase adjacent to its membrane anchor domain. J Biol Chem. 1989;264:20718–20722. - PubMed
-
- Brown DA, Crise B, Rose JK. Mechanism of membrane anchoring affects polarized expression of two proteins in MDCK cells. Science. 1989;245:1499–1501. - PubMed
-
- Casanova JE, Apodaca G, Mostov KE. An autonomous signal for basolateral sorting in the cytoplasmic domain of the polymeric immunoglobulin receptor. Cell. 1991;66:65–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

