p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum
- PMID: 9334338
- PMCID: PMC2139787
- DOI: 10.1083/jcb.139.2.327
p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum
Abstract
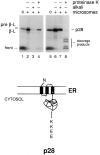
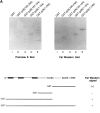
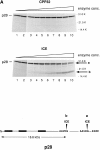
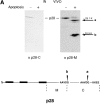
We have identified a human Bcl-2-interacting protein, p28 Bap31. It is a 28-kD (p28) polytopic integral protein of the endoplasmic reticulum whose COOH-terminal cytosolic region contains overlapping predicted leucine zipper and weak death effector homology domains, flanked on either side by identical caspase recognition sites. In cotransfected 293T cells, p28 is part of a complex that includes Bcl-2/Bcl-XL and procaspase-8 (pro-FLICE). Bax, a pro-apoptotic member of the Bcl-2 family, does not associate with the complex; however, it prevents Bcl-2 from doing so. In the absence (but not presence) of elevated Bcl-2 levels, apoptotic signaling by adenovirus E1A oncoproteins promote cleavage of p28 at the two caspase recognition sites. Purified caspase-8 (FLICE/MACH/Mch5) and caspase-1(ICE), but not caspase-3 (CPP32/apopain/ Yama), efficiently catalyze this reaction in vitro. The resulting NH2-terminal p20 fragment induces apoptosis when expressed ectopically in otherwise normal cells. Taken together, the results suggest that p28 Bap31 is part of a complex in the endoplasmic reticulum that mechanically bridges an apoptosis-initiating caspase, like procaspase-8, with the anti-apoptotic regulator Bcl-2 or Bcl-XL. This raises the possibility that the p28 complex contributes to the regulation of procaspase-8 or a related caspase in response to E1A, dependent on the status of the Bcl-2 setpoint within the complex.
Figures









Similar articles
-
Bcl-XL cooperatively associates with the Bap31 complex in the endoplasmic reticulum, dependent on procaspase-8 and Ced-4 adaptor.J Biol Chem. 1998 Feb 6;273(6):3140-3. doi: 10.1074/jbc.273.6.3140. J Biol Chem. 1998. PMID: 9452422
-
The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum.Proc Natl Acad Sci U S A. 2002 Apr 2;99(7):4331-6. doi: 10.1073/pnas.072088099. Epub 2002 Mar 26. Proc Natl Acad Sci U S A. 2002. PMID: 11917123 Free PMC article.
-
Evidence against a functional site for Bcl-2 downstream of caspase cascade in preventing apoptosis.Oncogene. 1997 Oct 16;15(16):1921-8. doi: 10.1038/sj.onc.1201370. Oncogene. 1997. PMID: 9365238
-
New insights in the role of Bcl-2 Bcl-2 and the endoplasmic reticulum.Apoptosis. 2002 Oct;7(5):441-7. doi: 10.1023/a:1020087108926. Apoptosis. 2002. PMID: 12207177 Review.
-
The endoplasmic reticulum as a site of protein degradation.Subcell Biochem. 1993;21:143-68. doi: 10.1007/978-1-4615-2912-5_7. Subcell Biochem. 1993. PMID: 8256264 Review. No abstract available.
Cited by
-
Functions of BCL-X L at the Interface between Cell Death and Metabolism.Int J Cell Biol. 2013;2013:705294. doi: 10.1155/2013/705294. Epub 2013 Feb 28. Int J Cell Biol. 2013. PMID: 23533418 Free PMC article.
-
Bcl-xL overexpression decreases GILZ levels and inhibits glucocorticoid-induced activation of caspase-8 and caspase-3 in mouse thymocytes.J Transl Autoimmun. 2020 Jan 28;3:100035. doi: 10.1016/j.jtauto.2020.100035. eCollection 2020. J Transl Autoimmun. 2020. PMID: 32803151 Free PMC article.
-
Contribution of apoptotic cell death to renal injury.J Cell Mol Med. 2001 Jan-Mar;5(1):18-32. doi: 10.1111/j.1582-4934.2001.tb00135.x. J Cell Mol Med. 2001. PMID: 12067448 Free PMC article. Review.
-
Intracellular cleavage of osteopontin by caspase-8 modulates hypoxia/reoxygenation cell death through p53.Proc Natl Acad Sci U S A. 2009 Sep 8;106(36):15326-31. doi: 10.1073/pnas.0903704106. Epub 2009 Aug 19. Proc Natl Acad Sci U S A. 2009. PMID: 19706414 Free PMC article.
-
Mechanisms of pre-apoptotic calreticulin exposure in immunogenic cell death.EMBO J. 2009 Mar 4;28(5):578-90. doi: 10.1038/emboj.2009.1. Epub 2009 Jan 22. EMBO J. 2009. PMID: 19165151 Free PMC article.
References
-
- Alnemri ES, Livingston DJ, Nicholson DW, Salvesen G, Thornberry NA, Wong WW, Yuan JY. Human ICE/CED-3 protease nomenclature. Cell. 1996;87:171. - PubMed
-
- Armstrong RC, Aja T, Xiang J, Gaur S, Krebs JF, Hoang K, Bai X, Korsmeyer SJ, Karanewsky DS, Fritz LC, Tomaselli KJ. Fas-induced activation of the cell death-related protease CPP32 is inhibited by Bcl-2 and by ICE family protease inhibitors. J Biol Chem. 1996;271:16850–16855. - PubMed
-
- Bauer MKA, Wesselborg S, Schulze-Osthoff K. The Caenorhabditis elegansdeath protein Ced-4 contains a motif with similarity to the mammalian death effector domain. FEBS Lett. 1997;402:256–258. - PubMed
-
- Blanar MA, Rutter WJ. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science (Wash DC) 1992;256:1014–1018. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

