Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration
- PMID: 9281581
- PMCID: PMC2136764
- DOI: 10.1083/jcb.138.5.1023
Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration
Abstract
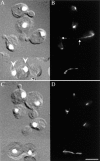
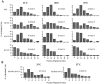
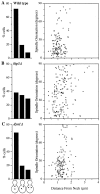
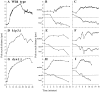
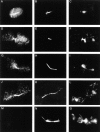
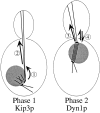
Spindle orientation and nuclear migration are crucial events in cell growth and differentiation of many eukaryotes. Here we show that KIP3, the sixth and final kinesin-related gene in Saccharomyces cerevisiae, is required for migration of the nucleus to the bud site in preparation for mitosis. The position of the nucleus in the cell and the orientation of the mitotic spindle was examined by microscopy of fixed cells and by time-lapse microscopy of individual live cells. Mutations in KIP3 and in the dynein heavy chain gene defined two distinct phases of nuclear migration: a KIP3-dependent movement of the nucleus toward the incipient bud site and a dynein-dependent translocation of the nucleus through the bud neck during anaphase. Loss of KIP3 function disrupts the unidirectional movement of the nucleus toward the bud and mitotic spindle orientation, causing large oscillations in nuclear position. The oscillatory motions sometimes brought the nucleus in close proximity to the bud neck, possibly accounting for the viability of a kip3 null mutant. The kip3 null mutant exhibits normal translocation of the nucleus through the neck and normal spindle pole separation kinetics during anaphase. Simultaneous loss of KIP3 and kinesin-related KAR3 function, or of KIP3 and dynein function, is lethal but does not block any additional detectable movement. This suggests that the lethality is due to the combination of sequential and possibly overlapping defects. Epitope-tagged Kip3p localizes to astral and central spindle microtubules and is also present throughout the cytoplasm and nucleus.
Figures



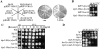

Comment in
-
Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle.J Cell Biol. 1997 Sep 8;138(5):957-60. doi: 10.1083/jcb.138.5.957. J Cell Biol. 1997. PMID: 9281575 Free PMC article. Review. No abstract available.
Similar articles
-
Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins.J Cell Biol. 1997 Sep 8;138(5):1041-53. doi: 10.1083/jcb.138.5.1041. J Cell Biol. 1997. PMID: 9281582 Free PMC article.
-
Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants.Mol Biol Cell. 2000 Nov;11(11):3949-61. doi: 10.1091/mbc.11.11.3949. Mol Biol Cell. 2000. PMID: 11071919 Free PMC article.
-
The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle.J Cell Biol. 1994 Apr;125(1):143-58. doi: 10.1083/jcb.125.1.143. J Cell Biol. 1994. PMID: 8138567 Free PMC article.
-
Mitotic motors in Saccharomyces cerevisiae.Biochim Biophys Acta. 2000 Mar 17;1496(1):99-116. doi: 10.1016/s0167-4889(00)00012-4. Biochim Biophys Acta. 2000. PMID: 10722880 Review.
-
Motoring to the finish: kinesin and dynein work together to orient the yeast mitotic spindle.J Cell Biol. 1997 Sep 8;138(5):957-60. doi: 10.1083/jcb.138.5.957. J Cell Biol. 1997. PMID: 9281575 Free PMC article. Review. No abstract available.
Cited by
-
Role and regulation of kinesin-8 motors through the cell cycle.Syst Synth Biol. 2014 Sep;8(3):205-13. doi: 10.1007/s11693-014-9140-z. Epub 2014 Mar 23. Syst Synth Biol. 2014. PMID: 25136382 Free PMC article.
-
Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase.Genetics. 2006 Jul;173(3):1187-96. doi: 10.1534/genetics.105.050062. Epub 2006 Apr 19. Genetics. 2006. PMID: 16624923 Free PMC article.
-
Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase.Cell Cycle. 2010 Jul 1;9(13):2581-8. doi: 10.4161/cc.9.13.12076. Cell Cycle. 2010. PMID: 20603597 Free PMC article.
-
Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility.PLoS One. 2009;4(3):e4854. doi: 10.1371/journal.pone.0004854. Epub 2009 Mar 16. PLoS One. 2009. PMID: 19287503 Free PMC article.
-
Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins.J Cell Biol. 1997 Sep 8;138(5):1041-53. doi: 10.1083/jcb.138.5.1041. J Cell Biol. 1997. PMID: 9281582 Free PMC article.
References
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

