Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function
- PMID: 9265645
- PMCID: PMC2138052
- DOI: 10.1083/jcb.138.4.771
Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function
Abstract
Cofilin is an actin depolymerizing protein found widely distributed in animals and plants. We have used electron cryomicroscopy and helical reconstruction to identify its binding site on actin filaments. Cofilin binds filamentous (F)-actin cooperatively by bridging two longitudinally associated actin subunits. The binding site is centered axially at subdomain 2 of the lower actin subunit and radially at the cleft between subdomains 1 and 3 of the upper actin subunit. Our work has revealed a totally unexpected (and unique) property of cofilin, namely, its ability to change filament twist. As a consequence of this change in twist, filaments decorated with cofilin have much shorter 'actin crossovers' ( approximately 75% of those normally observed in F-actin structures). Although their binding sites are distinct, cofilin and phalloidin do not bind simultaneously to F-actin. This is the first demonstration of a protein that excludes another actin-binding molecule by changing filament twist. Alteration of F-actin structure by cofilin/ADF appears to be a novel mechanism through which the actin cytoskeleton may be regulated or remodeled.
Figures


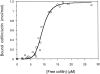

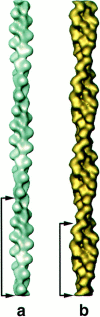


| 0.8515 | 0.5072 | 0.1331 | |||
| −0.3293 | 0.7147 | −0.6170 | |||
| −0.4081 | 0.4815 | 0.7756 | |||
| −1.7062 | −12.3144 | −12.5306 |
Similar articles
-
Uncoupling actin filament fragmentation by cofilin from increased subunit turnover.J Mol Biol. 2000 May 12;298(4):649-61. doi: 10.1006/jmbi.2000.3688. J Mol Biol. 2000. PMID: 10788327
-
Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility.J Cell Biol. 1997 Mar 24;136(6):1307-22. doi: 10.1083/jcb.136.6.1307. J Cell Biol. 1997. PMID: 9087445 Free PMC article.
-
ADF/cofilin weakens lateral contacts in the actin filament.J Mol Biol. 1999 Aug 20;291(3):513-9. doi: 10.1006/jmbi.1999.2968. J Mol Biol. 1999. PMID: 10448032 Review.
-
The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics.Mol Biol Cell. 1995 Nov;6(11):1423-31. doi: 10.1091/mbc.6.11.1423. Mol Biol Cell. 1995. PMID: 8589446 Free PMC article. Review. No abstract available.
-
Actin hydrophobic loop 262-274 and filament nucleation and elongation.J Mol Biol. 2008 Jan 18;375(3):793-801. doi: 10.1016/j.jmb.2007.10.076. Epub 2007 Nov 4. J Mol Biol. 2008. PMID: 18037437 Free PMC article.
Cited by
-
ADF/cofilin is not essential but is critically important for actin activities during phagocytosis in Tetrahymena thermophila.Eukaryot Cell. 2013 Aug;12(8):1080-6. doi: 10.1128/EC.00074-13. Epub 2013 May 31. Eukaryot Cell. 2013. PMID: 23729382 Free PMC article.
-
Troponin and a Myopathy-Linked Mutation in TPM3 Inhibit Cofilin-2-Induced Thin Filament Depolymerization.Int J Mol Sci. 2023 Nov 17;24(22):16457. doi: 10.3390/ijms242216457. Int J Mol Sci. 2023. PMID: 38003645 Free PMC article.
-
Mechanosensing through Direct Binding of Tensed F-Actin by LIM Domains.Dev Cell. 2020 Nov 23;55(4):468-482.e7. doi: 10.1016/j.devcel.2020.09.022. Epub 2020 Oct 14. Dev Cell. 2020. PMID: 33058779 Free PMC article.
-
Regulation of pulmonary endothelial barrier function by kinases.Am J Physiol Lung Cell Mol Physiol. 2016 Nov 1;311(5):L832-L845. doi: 10.1152/ajplung.00233.2016. Epub 2016 Sep 23. Am J Physiol Lung Cell Mol Physiol. 2016. PMID: 27663990 Free PMC article. Review.
-
Actin mediates the nanoscale membrane organization of the clustered membrane protein influenza hemagglutinin.Biophys J. 2013 May 21;104(10):2182-92. doi: 10.1016/j.bpj.2013.03.054. Biophys J. 2013. PMID: 23708358 Free PMC article.
References
-
- Aebi U, Millonig R, Salvo H, Engel A. The three-dimensional structure of the actin filament revisited. Ann NY Acad Sci. 1986;483:100–119. - PubMed
-
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. - PubMed
-
- Amos LA. Combination of data from helical particles: correlation and selection. J Mol Biol. 1975;99:65–73. - PubMed
-
- Bamburg JR, Harris HE, Weeds AG. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS (Fed Eur Biochem Soc) Lett. 1980;121:178–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

