Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination
- PMID: 9256462
- PMCID: PMC23120
- DOI: 10.1073/pnas.94.17.9214
Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination
Abstract
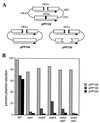
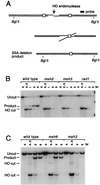
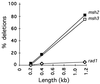
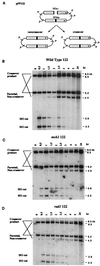
When gene conversion is initiated by a double-strand break (DSB), any nonhomologous DNA that may be present at the ends must be removed before new DNA synthesis can be initiated. In Saccharomyces cerevisiae, removal of nonhomologous ends depends not only on the nucleotide excision repair endonuclease Rad1/Rad10 but also on Msh2 and Msh3, two proteins that are required to correct mismatched bp. These proteins have no effect when DSB ends are homologous to the donor, either in the kinetics of recombination or in the proportion of gene conversions associated with crossing-over. A second DSB repair pathway, single-strand annealing also requires Rad1/Rad10 and Msh2/Msh3, but reveals a difference in their roles. When the flanking homologous regions that anneal are 205 bp, the requirement for Msh2/Msh3 is as great as for Rad1/Rad10; but when the annealing partners are 1,170 bp, Msh2/Msh3 have little effect, while Rad1/Rad10 are still required. Mismatch repair proteins Msh6, Pms1, and Mlh1 are not required. We suggest Msh2 and Msh3 recognize not only heteroduplex loops and mismatched bp, but also branched DNA structures with a free 3' tail.
Figures

Similar articles
-
Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae.Genetics. 1996 Mar;142(3):727-36. doi: 10.1093/genetics/142.3.727. Genetics. 1996. PMID: 8849883 Free PMC article.
-
Coordination of Rad1-Rad10 interactions with Msh2-Msh3, Saw1 and RPA is essential for functional 3' non-homologous tail removal.Nucleic Acids Res. 2018 Jun 1;46(10):5075-5096. doi: 10.1093/nar/gky254. Nucleic Acids Res. 2018. PMID: 29660012 Free PMC article.
-
Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae.Mol Cell Biol. 1997 Nov;17(11):6765-71. doi: 10.1128/MCB.17.11.6765. Mol Cell Biol. 1997. PMID: 9343441 Free PMC article.
-
A tale of tails: insights into the coordination of 3' end processing during homologous recombination.Bioessays. 2009 Mar;31(3):315-21. doi: 10.1002/bies.200800195. Bioessays. 2009. PMID: 19260026 Free PMC article. Review.
-
Genomic amplification of the human DHFR/MSH3 locus remodels mismatch recognition and repair activities.Adv Enzyme Regul. 1999;39:129-41. doi: 10.1016/s0065-2571(98)00013-2. Adv Enzyme Regul. 1999. PMID: 10470370 Review.
Cited by
-
Assessment of anti-recombination and double-strand break-induced gene conversion in human cells by a chromosomal reporter.J Biol Chem. 2012 Aug 24;287(35):29543-53. doi: 10.1074/jbc.M112.352302. Epub 2012 Jul 7. J Biol Chem. 2012. PMID: 22773873 Free PMC article.
-
MutSβ protects common fragile sites by facilitating homology-directed repair at DNA double-strand breaks with secondary structures.Nucleic Acids Res. 2024 Feb 9;52(3):1120-1135. doi: 10.1093/nar/gkad1112. Nucleic Acids Res. 2024. PMID: 38038265 Free PMC article.
-
DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase.Mol Cell Biol. 2005 Mar;25(6):2297-309. doi: 10.1128/MCB.25.6.2297-2309.2005. Mol Cell Biol. 2005. PMID: 15743825 Free PMC article.
-
Msh2 blocks an alternative mechanism for non-homologous tail removal during single-strand annealing in Saccharomyces cerevisiae.PLoS One. 2009 Oct 16;4(10):e7488. doi: 10.1371/journal.pone.0007488. PLoS One. 2009. PMID: 19834615 Free PMC article.
-
Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination.Nucleic Acids Res. 2004 Oct 5;32(17):5239-48. doi: 10.1093/nar/gkh822. Print 2004. Nucleic Acids Res. 2004. PMID: 15466591 Free PMC article.
References
-
- Weng Y S, Whelden J, Gunn L, Nickoloff J A. Curr Genet. 1996;29:335–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

