Regulation of calreticulin gene expression by calcium
- PMID: 9245785
- PMCID: PMC2141645
- DOI: 10.1083/jcb.138.3.547
Regulation of calreticulin gene expression by calcium
Abstract
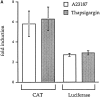
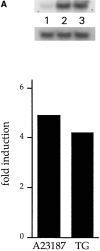
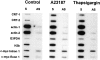
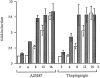
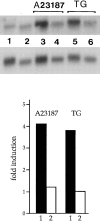
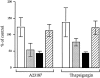
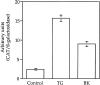
We have isolated and characterized a 12-kb mouse genomic DNA fragment containing the entire calreticulin gene and 2.14 kb of the promoter region. The mouse calreticulin gene consists of nine exons and eight introns, and it spans 4.2 kb of genomic DNA. A 1.8-kb fragment of the calreticulin promoter was subcloned into a reporter gene plasmid containing chloramphenicol acetyltransferase. This construct was then used in transient and stable transfection of NIH/ 3T3 cells. Treatment of transfected cells either with the Ca2+ ionophore A23187, or with the ER Ca2+-ATPase inhibitor thapsigargin, resulted in a five- to sevenfold increase of the expression of chloramphenicol acetyltransferase protein. Transactivation of the calreticulin promoter was also increased by fourfold in NIH/3T3 cells treated with bradykinin, a hormone that induces Ca2+ release from the intracellular Ca2+ stores. Analysis of the promoter deletion constructs revealed that A23187- and thapsigargin-responsive regions are confined to two regions (-115 to -260 and -685 to -1,763) in the calreticulin promoter that contain the CCAAT nucleotide sequences. Northern blot analysis of cells treated with A23187, or with thapsigargin, revealed a fivefold increase in calreticulin mRNA levels. Thapsigargin also induced a fourfold increase in calreticulun protein levels. Importantly, we show by nuclear run-on transcription analysis that calreticulin gene transcription is increased in NIH/3T3 cells treated with A23187 and thapsigargin in vivo. This increase in gene expression required over 4 h of continuous incubation with the drugs and was also sensitive to treatment with cycloheximide, suggesting that it is dependent on protein synthesis. Changes in the concentration of extracellular and cytoplasmic Ca2+ did not affect the increased expression of the calreticulin gene. These studies suggest that stress response to the depletion of intracellular Ca2+ stores induces expression of the calreticulin gene in vitro and in vivo.
Figures



Similar articles
-
Induction of calreticulin expression in HeLa cells by depletion of the endoplasmic reticulum Ca2+ store and inhibition of N-linked glycosylation.Biochem J. 1996 Sep 1;318 ( Pt 2)(Pt 2):555-60. doi: 10.1042/bj3180555. Biochem J. 1996. PMID: 8809046 Free PMC article.
-
Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca(2+)-ATPase inhibitor thapsigargin.J Biol Chem. 1993 Jun 5;268(16):12003-9. J Biol Chem. 1993. PMID: 8505325
-
Functional specialization of calreticulin domains.J Cell Biol. 2001 Sep 3;154(5):961-72. doi: 10.1083/jcb.200102073. Epub 2001 Aug 27. J Cell Biol. 2001. PMID: 11524434 Free PMC article.
-
Calreticulin: one protein, one gene, many functions.Biochem J. 1999 Dec 1;344 Pt 2(Pt 2):281-92. Biochem J. 1999. PMID: 10567207 Free PMC article. Review.
-
Endoplasmic reticulum in the heart, a forgotten organelle?Mol Cell Biochem. 2001 Sep;225(1-):1-6. doi: 10.1023/a:1012209923231. Mol Cell Biochem. 2001. PMID: 11716351 Review.
Cited by
-
Transgenic mouse and cell culture models demonstrate a lack of mechanistic connection between endoplasmic reticulum stress and tau dysfunction.J Neurosci Res. 2010 Jul;88(9):1951-61. doi: 10.1002/jnr.22359. J Neurosci Res. 2010. PMID: 20143409 Free PMC article.
-
Metallothionein alleviates oxidative stress-induced endoplasmic reticulum stress and myocardial dysfunction.J Mol Cell Cardiol. 2009 Aug;47(2):228-37. doi: 10.1016/j.yjmcc.2009.03.018. Epub 2009 Apr 1. J Mol Cell Cardiol. 2009. PMID: 19344729 Free PMC article.
-
Unfolded protein response-induced dysregulation of calcium homeostasis promotes retinal degeneration in rat models of autosomal dominant retinitis pigmentosa.Cell Death Dis. 2016 Feb 4;7(2):e2085. doi: 10.1038/cddis.2015.325. Cell Death Dis. 2016. PMID: 26844699 Free PMC article.
-
TMEM203 Is a Novel Regulator of Intracellular Calcium Homeostasis and Is Required for Spermatogenesis.PLoS One. 2015 May 21;10(5):e0127480. doi: 10.1371/journal.pone.0127480. eCollection 2015. PLoS One. 2015. PMID: 25996873 Free PMC article.
-
High-throughput screening discovers antifibrotic properties of haloperidol by hindering myofibroblast activation.JCI Insight. 2019 Apr 18;4(8):e123987. doi: 10.1172/jci.insight.123987. eCollection 2019 Apr 18. JCI Insight. 2019. PMID: 30996132 Free PMC article.
References
-
- Ausubel, F.M., R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1989. Current Protocols in Molecular Biology. Vols. 1 and 2.
-
- Bachs O, Agell N, Carafoli E. Calcium and calmodulin function in the cell nucleus. Biochim Biophys Acta. 1992;1113:259–270. - PubMed
-
- Baksh S, Burns K, Andrin C, Michalak M. Interaction of calreticulin with protein disulfide isomerase. J Biol Chem. 1995;270:31338–31344. - PubMed
-
- Bastianutto C, Clementi E, Codazzi F, Podini P, De Giorgi F, Rizzuto R, Meldolesi J, Pozzan T. Overexpression of calreticulin increases the Ca2+ capacity of rapidly exchanging Ca2+stores and reveals aspects of their lumenal microenvironment and function. J Cell Biol. 1995;130:847–855. - PMC - PubMed
-
- Bergeron JGM, Brenner MB, Thomas DY, Williams D. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. TIBS. 1994;19:124–128. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

