Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism
- PMID: 9214379
- PMCID: PMC2139945
- DOI: 10.1083/jcb.138.1.37
Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism
Abstract
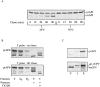
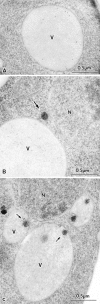
The yeast vacuolar protein aminopeptidase I (API) is synthesized as a cytosolic precursor that is transported to the vacuole by a nonclassical targeting mechanism. Recent genetic studies indicate that the biosynthetic pathway that transports API uses many of the same molecular components as the degradative autophagy pathway. This overlap coupled with both in vitro and in vivo analysis of API import suggested that, like autophagy, API transport is vesicular. Subcellular fractionation experiments demonstrate that API precursor (prAPI) initially enters a nonvacuolar cytosolic compartment. In addition, subvacuolar vesicles containing prAPI were purified from a mutant strain defective in breakdown of autophagosomes, further indicating that prAPI enters the vacuole inside a vesicle. The purified subvacuolar vesicles do not appear to contain vacuolar marker proteins. Immunogold EM confirms that prAPI is localized in cytosolic and in subvacuolar vesicles in a mutant strain defective in autophagic body degradation. These data suggest that cytosolic vesicles containing prAPI fuse with the vacuole to release a membrane-bounded intermediate compartment that is subsequently broken down, allowing API maturation.
Figures


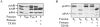
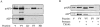
Similar articles
-
Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole.J Cell Biol. 2001 Apr 16;153(2):381-96. doi: 10.1083/jcb.153.2.381. J Cell Biol. 2001. PMID: 11309418 Free PMC article.
-
Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome.J Cell Biol. 1997 Dec 29;139(7):1687-95. doi: 10.1083/jcb.139.7.1687. J Cell Biol. 1997. PMID: 9412464 Free PMC article.
-
Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p.EMBO J. 1999 Nov 1;18(21):6005-16. doi: 10.1093/emboj/18.21.6005. EMBO J. 1999. PMID: 10545112 Free PMC article.
-
Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review.
-
Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells.Annu Rev Biochem. 2000;69:303-42. doi: 10.1146/annurev.biochem.69.1.303. Annu Rev Biochem. 2000. PMID: 10966461 Review.
Cited by
-
The Cytoplasm-to-Vacuole Targeting Pathway: A Historical Perspective.Int J Cell Biol. 2012;2012:142634. doi: 10.1155/2012/142634. Epub 2012 Feb 20. Int J Cell Biol. 2012. PMID: 22481942 Free PMC article.
-
The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article.
-
Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway.J Biol Chem. 2010 Sep 17;285(38):29599-607. doi: 10.1074/jbc.M110.113670. Epub 2010 Jul 8. J Biol Chem. 2010. PMID: 20615880 Free PMC article.
-
The machinery of macroautophagy.Cell Res. 2014 Jan;24(1):24-41. doi: 10.1038/cr.2013.168. Epub 2013 Dec 24. Cell Res. 2014. PMID: 24366339 Free PMC article. Review.
-
Assays to Monitor Autophagy in Saccharomyces cerevisiae.Cells. 2017 Jul 13;6(3):23. doi: 10.3390/cells6030023. Cells. 2017. PMID: 28703742 Free PMC article. Review.
References
-
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. - PubMed
-
- Baker D, Schekman R. Reconstitution of protein transport using broken yeast spheroplasts. Methods Cell Biol. 1989;31:127–141. - PubMed
-
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
-
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell. 1995;83:513–516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

