Prion protein NMR structure and species barrier for prion diseases
- PMID: 9207082
- PMCID: PMC23812
- DOI: 10.1073/pnas.94.14.7281
Prion protein NMR structure and species barrier for prion diseases
Abstract
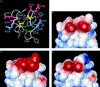
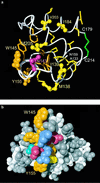
The structural basis of species specificity of transmissible spongiform encephalopathies, such as bovine spongiform encephalopathy or "mad cow disease" and Creutzfeldt-Jakob disease in humans, has been investigated using the refined NMR structure of the C-terminal domain of the mouse prion protein with residues 121-231. A database search for mammalian prion proteins yielded 23 different sequences for the fragment 124-226, which display a high degree of sequence identity and show relevant amino acid substitutions in only 18 of the 103 positions. Except for a unique isolated negative surface charge in the bovine protein, the amino acid differences are clustered in three distinct regions of the three-dimensional structure of the cellular form of the prion protein. Two of these regions represent potential species-dependent surface recognition sites for protein-protein interactions, which have independently been implicated from in vitro and in vivo studies of prion protein transformation. The third region consists of a cluster of interior hydrophobic side chains that may affect prion protein transformation at later stages, after initial conformational changes in the cellular protein.
Figures
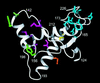

Similar articles
-
Toward the molecular basis of inherited prion diseases: NMR structure of the human prion protein with V210I mutation.J Mol Biol. 2011 Sep 30;412(4):660-73. doi: 10.1016/j.jmb.2011.07.067. Epub 2011 Aug 4. J Mol Biol. 2011. PMID: 21839748
-
Molecular dynamics studies on the NMR and X-ray structures of rabbit prion proteins.J Theor Biol. 2014 Feb 7;342:70-82. doi: 10.1016/j.jtbi.2013.10.005. Epub 2013 Oct 31. J Theor Biol. 2014. PMID: 24184221
-
Molecular dynamics studies on 3D structures of the hydrophobic region PrP(109-136).Acta Biochim Biophys Sin (Shanghai). 2013 Jun;45(6):509-19. doi: 10.1093/abbs/gmt031. Epub 2013 Apr 5. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23563221 Review.
-
Prions and prion diseases: fundamentals and mechanistic details.J Microbiol Biotechnol. 2007 Jul;17(7):1059-70. J Microbiol Biotechnol. 2007. PMID: 18051314 Review.
-
Transmissible spongiform encephalopathies.Biochem Biophys Res Commun. 1998 Sep 18;250(2):187-93. doi: 10.1006/bbrc.1998.9169. Biochem Biophys Res Commun. 1998. PMID: 9753605 Review.
Cited by
-
Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state.EMBO J. 1999 Jun 15;18(12):3193-203. doi: 10.1093/emboj/18.12.3193. EMBO J. 1999. PMID: 10369660 Free PMC article.
-
A molecular switch controls interspecies prion disease transmission in mice.J Clin Invest. 2010 Jul;120(7):2590-9. doi: 10.1172/JCI42051. Epub 2010 Jun 14. J Clin Invest. 2010. PMID: 20551516 Free PMC article.
-
Normal prion protein has an activity like that of superoxide dismutase.Biochem J. 1999 Nov 15;344 Pt 1(Pt 1):1-5. Biochem J. 1999. PMID: 10548526 Free PMC article.
-
Refinement of under-determined loops of Human Prion Protein by database-derived distance constraints.Int J Data Min Bioinform. 2009;3(4):454-68. doi: 10.1504/ijdmb.2009.029206. Int J Data Min Bioinform. 2009. PMID: 20052907 Free PMC article.
-
NMR solution structure of the human prion protein.Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):145-50. doi: 10.1073/pnas.97.1.145. Proc Natl Acad Sci U S A. 2000. PMID: 10618385 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

