Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding
- PMID: 9207072
- PMCID: PMC23798
- DOI: 10.1073/pnas.94.14.7221
Slow dimer dissociation of the TATA binding protein dictates the kinetics of DNA binding
Abstract
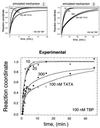
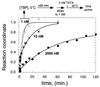
The association of the TATA binding protein (TBP) to eukaryotic promoters is a possible rate-limiting step in gene expression. Slow promoter binding might be related to TBP's ability to occlude its DNA binding domain through dimerization. Using a "pull-down" based assay, we find that TBP dimers dissociate slowly (t1/2 = 6-10 min), and thus present a formidable kinetic barrier to TATA binding. At 10 nM, TBP appears to exist as a mixed population of monomers and dimers. In this state, TATA binding displays burst kinetics that appears to reflect rapid binding of monomers and slow dissociation of dimers. The kinetics of the slow phase is in excellent agreement with direct measurements of the kinetics of dimer dissociation.
Figures


Similar articles
-
Dimer dissociation and thermosensitivity kinetics of the Saccharomyces cerevisiae and human TATA binding proteins.Biochemistry. 1999 Aug 31;38(35):11340-8. doi: 10.1021/bi990911p. Biochemistry. 1999. PMID: 10471284
-
TFIIA regulates TBP and TFIID dimers.Mol Cell. 1999 Sep;4(3):451-7. doi: 10.1016/s1097-2765(00)80453-0. Mol Cell. 1999. PMID: 10518227
-
Dimerization of the TATA binding protein.J Biol Chem. 1995 Jun 9;270(23):13842-9. doi: 10.1074/jbc.270.23.13842. J Biol Chem. 1995. PMID: 7775442
-
Dynamic interplay of TFIIA, TBP and TATA DNA.J Mol Biol. 1997 Aug 8;271(1):61-75. doi: 10.1006/jmbi.1997.1152. J Mol Biol. 1997. PMID: 9300055
-
X-ray crystallographic studies of eukaryotic transcription initiation factors.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):483-9. doi: 10.1098/rstb.1996.0046. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735270 Review.
Cited by
-
Structural and functional analysis of mutations along the crystallographic dimer interface of the yeast TATA binding protein.Mol Cell Biol. 2003 May;23(9):3186-201. doi: 10.1128/MCB.23.9.3186-3201.2003. Mol Cell Biol. 2003. PMID: 12697819 Free PMC article.
-
A kinetic model of TBP auto-regulation exhibits bistability.Biol Direct. 2010 Aug 5;5:50. doi: 10.1186/1745-6150-5-50. Biol Direct. 2010. PMID: 20687914 Free PMC article.
-
A single point mutation in TFIIA suppresses NC2 requirement in vivo.EMBO J. 2000 Feb 15;19(4):672-82. doi: 10.1093/emboj/19.4.672. EMBO J. 2000. PMID: 10675336 Free PMC article.
-
Transcription factors that influence RNA polymerases I and II: To what extent is mechanism of action conserved?Biochim Biophys Acta Gene Regul Mech. 2017 Feb;1860(2):246-255. doi: 10.1016/j.bbagrm.2016.10.010. Epub 2016 Oct 27. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27989933 Free PMC article. Review.
-
Analysis of homodimer formation in 12-oxophytodienoate reductase 3 in solutio and crystallo challenges the physiological role of the dimer.Sci Rep. 2024 Aug 5;14(1):18093. doi: 10.1038/s41598-024-69160-6. Sci Rep. 2024. PMID: 39103552 Free PMC article.
References
-
- Hernandez N. Genes Dev. 1993;7:1291–1308. - PubMed
-
- Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. - PubMed
-
- Pugh B F. Curr Opin Cell Biol. 1996;8:303–311. - PubMed
-
- Wang W, Gralla J D, Carey M. Genes Dev. 1992;6:1716–1727. - PubMed
-
- Lieberman P M, Berk A J. Genes Dev. 1994;8:995–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

