Transient aggregates in protein folding are easily mistaken for folding intermediates
- PMID: 9177173
- PMCID: PMC21005
- DOI: 10.1073/pnas.94.12.6084
Transient aggregates in protein folding are easily mistaken for folding intermediates
Abstract
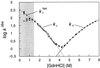
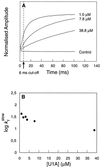
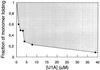
It has been questioned recently whether populated intermediates are important for the protein folding process or are artefacts trapped in nonproductive pathways. We report here that the rapidly formed intermediate of the spliceosomal protein U1A is an off-pathway artefact caused by transient aggregation of denatured protein under native conditions. Transient aggregates are easily mistaken for structured monomers and could be a general problem in time-resolved folding studies.
Figures
Similar articles
-
Transient aggregation and stable dimerization induced by introducing an Alzheimer sequence into a water-soluble protein.Biochemistry. 2004 Oct 19;43(41):12964-78. doi: 10.1021/bi048509k. Biochemistry. 2004. PMID: 15476390
-
High-energy channeling in protein folding.Biochemistry. 1997 Jun 24;36(25):7633-7. doi: 10.1021/bi970210x. Biochemistry. 1997. PMID: 9201903
-
Formation of short-lived protein aggregates directly from the coil in two-state folding.Biochemistry. 1999 Oct 5;38(40):13006-12. doi: 10.1021/bi9909997. Biochemistry. 1999. PMID: 10529170
-
Spliceosomal UsnRNP biogenesis, structure and function.Curr Opin Cell Biol. 2001 Jun;13(3):290-301. doi: 10.1016/s0955-0674(00)00211-8. Curr Opin Cell Biol. 2001. PMID: 11343899 Review.
-
Architecture of the spliceosome.Biochemistry. 2012 Apr 24;51(16):3321-33. doi: 10.1021/bi201215r. Epub 2012 Apr 10. Biochemistry. 2012. PMID: 22471593 Review.
Cited by
-
The sequences of small proteins are not extensively optimized for rapid folding by natural selection.Proc Natl Acad Sci U S A. 1998 Apr 28;95(9):4982-6. doi: 10.1073/pnas.95.9.4982. Proc Natl Acad Sci U S A. 1998. PMID: 9560214 Free PMC article.
-
A protein folding pathway with multiple folding intermediates at atomic resolution.Proc Natl Acad Sci U S A. 2005 Apr 5;102(14):5026-31. doi: 10.1073/pnas.0501372102. Epub 2005 Mar 25. Proc Natl Acad Sci U S A. 2005. PMID: 15793003 Free PMC article.
-
Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers.Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15893-8. doi: 10.1073/pnas.0403979101. Epub 2004 Nov 2. Proc Natl Acad Sci U S A. 2004. PMID: 15522970 Free PMC article.
-
Competition of individual domain folding with inter-domain interaction in WW domain engineered repeat proteins.Phys Chem Chem Phys. 2019 Nov 13;21(44):24393-24405. doi: 10.1039/c8cp07775d. Phys Chem Chem Phys. 2019. PMID: 31663524 Free PMC article.
-
Observation of strange kinetics in protein folding.Proc Natl Acad Sci U S A. 1999 May 25;96(11):6031-6. doi: 10.1073/pnas.96.11.6031. Proc Natl Acad Sci U S A. 1999. PMID: 10339536 Free PMC article.
References
-
- Udgaonkar J B, Baldwin R L. Nature (London) 1988;335:700–704. - PubMed
-
- Matouschek A, Kellis J T, Serrano L, Bycroft M, Fersht A R. Nature (London) 1990;346:440–445. - PubMed
-
- Kim P S, Baldwin R L. Annu Rev Biochem. 1990;59:631–660. - PubMed
-
- Radford S E, Dobson C M, Evans P A. Nature (London) 1992;358:302–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

