Targeted expression of major histocompatibility complex (MHC) class II molecules demonstrates that dendritic cells can induce negative but not positive selection of thymocytes in vivo
- PMID: 9053454
- PMCID: PMC2196043
- DOI: 10.1084/jem.185.3.541
Targeted expression of major histocompatibility complex (MHC) class II molecules demonstrates that dendritic cells can induce negative but not positive selection of thymocytes in vivo
Abstract
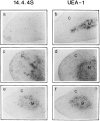
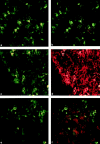
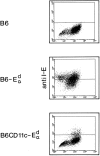
It is well established that lymphoid dendritic cells (DC) play an important role in the immune system. Beside their role as potent inducers of primary T cell responses, DC seem to play a crucial part as major histocompatibility complex (MHC) class II+ "interdigitating cells" in the thymus during thymocyte development. Thymic DC have been implicated in tolerance induction and also by some authors in inducing major histocompatibility complex restriction of thymocytes. Most of our knowledge about thymic DC was obtained using highly invasive and manipulatory experimental protocols such as thymus reaggregation cultures, suspension cultures, thymus grafting, and bone marrow reconstitution experiments. The DC used in those studies had to go through extensive isolation procedures or were cultured with recombinant growth factors. Since the functions of DC after these in vitro manipulations have been reported to be not identical to those of DC in vivo, we intended to establish a system that would allow us to investigate DC function avoiding artificial interferences due to handling. Here we present a transgenic mouse model in which we targeted gene expression specifically to DC. Using the CD 11c promoter we expressed MHC class II I-E molecules specifically on DC of all tissues, but not on other cell types. We report that I-E expression on thymic DC is sufficient to negatively select I-E reactive CD4+ T cells, and to a less complete extent, CD8+ T cells. In contrast, it only DC expressed I-E in a class II-deficient background, positive selection of CD4+ T cells could not be observed. Thus negative, but not positive, selection events can be induced by DC in vivo.
Figures



Similar articles
-
Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells.J Exp Med. 1997 Oct 20;186(8):1223-32. doi: 10.1084/jem.186.8.1223. J Exp Med. 1997. PMID: 9334361 Free PMC article.
-
The role of dendritic cells in T cell selection and survival.J Leukoc Biol. 1999 Aug;66(2):331-5. doi: 10.1002/jlb.66.2.331. J Leukoc Biol. 1999. PMID: 10449177
-
Inhibition of I-Ad-, but not Db-restricted peptide-induced thymic apoptosis by glucocorticoid receptor antagonist RU486 in T cell receptor transgenic mice.Eur J Immunol. 1996 Feb;26(2):428-34. doi: 10.1002/eji.1830260224. Eur J Immunol. 1996. PMID: 8617314
-
Thymic dendritic cells: phenotype and function.Int Rev Immunol. 1990;6(2-3):187-96. doi: 10.3109/08830189009056629. Int Rev Immunol. 1990. PMID: 2152502 Review.
-
Investigating central tolerance with reaggregate thymus organ cultures.Methods Mol Biol. 2007;380:185-96. doi: 10.1007/978-1-59745-395-0_11. Methods Mol Biol. 2007. PMID: 17876094 Review.
Cited by
-
Legionella pneumophila infection up-regulates dendritic cell Toll-like receptor 2 (TLR2)/TLR4 expression and key maturation markers.Infect Immun. 2007 Jun;75(6):3205-8. doi: 10.1128/IAI.01950-06. Epub 2007 Mar 19. Infect Immun. 2007. PMID: 17371856 Free PMC article.
-
Induction of polyomavirus-specific CD8(+) T lymphocytes by distinct dendritic cell subpopulations.J Virol. 2001 Jan;75(1):544-7. doi: 10.1128/JVI.75.1.544-547.2001. J Virol. 2001. PMID: 11119625 Free PMC article.
-
Constitutive TL1A (TNFSF15) expression on lymphoid or myeloid cells leads to mild intestinal inflammation and fibrosis.PLoS One. 2011 Jan 11;6(1):e16090. doi: 10.1371/journal.pone.0016090. PLoS One. 2011. PMID: 21264313 Free PMC article.
-
The Importance of Dendritic Cells in Maintaining Immune Tolerance.J Immunol. 2017 Mar 15;198(6):2223-2231. doi: 10.4049/jimmunol.1601629. J Immunol. 2017. PMID: 28264998 Free PMC article. Review.
-
Chronic CD27-CD70 costimulation promotes type 1-specific polarization of effector Tregs.Front Immunol. 2023 Mar 13;14:1023064. doi: 10.3389/fimmu.2023.1023064. eCollection 2023. Front Immunol. 2023. PMID: 36993956 Free PMC article.
References
-
- Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. - PubMed
-
- Nossal GJ. Negative selection of lymphocytes. Cell. 1994;76:229–239. - PubMed
-
- Anderson G, Moore NC, Owen JJT, Jenkinson EJ. Cellular interactions in thymocyte development. Annu Rev Immunol. 1996;14:73–99. - PubMed
-
- Pircher H, Brduscha K, Steinhoff U, Kasai M, Mizuochi T, Zinkernagel RM, Hengartner H, Kyewski B, Muller KP. Tolerance induction by clonal deletion of CD4+8+ thymocytes in vitro does not require dedicated antigen-presenting cells. Eur J Immunol. 1993;23:669–674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

