The seizure locus encodes the Drosophila homolog of the HERG potassium channel
- PMID: 8994043
- PMCID: PMC6573183
- DOI: 10.1523/JNEUROSCI.17-03-00882.1997
The seizure locus encodes the Drosophila homolog of the HERG potassium channel
Abstract
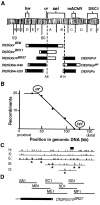
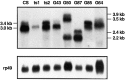
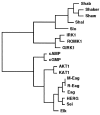
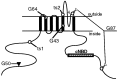
Mutations in the seizure (sei) locus cause temperature-induced hyperactivity, followed by paralysis. Gene cloning studies have established that the seizure gene product is the Drosophila homolog of HERG, a member of the eag family of K+ channels implicated in one form of hereditary long QT syndrome in humans. A series of five null alleles with premature stop codons are all recessive, but viable. A missense mutation in the sei gene, which changes the charge at a conserved glutamate residue near the outer mouth of the pore, has a semidominant phenotype, suggesting that the mutant seizure protein acts as a poison in a multimeric complex. Transformation rescue of a null allele with a cDNA under the control of an inducible promoter demonstrates that induced expression of seizure potassium channels in adults rescues the paralytic phenotype. This rescue decays with a t1/2 of approximately 1-1.5 d after gene induction is discontinued, providing the first estimate of ion channel stability in an intact, multicellular animal.
Figures

Similar articles
-
The eag family of K+ channels in Drosophila and mammals.Ann N Y Acad Sci. 1999 Apr 30;868:356-69. doi: 10.1111/j.1749-6632.1999.tb11297.x. Ann N Y Acad Sci. 1999. PMID: 10414305
-
The Drosophila erg K+ channel polypeptide is encoded by the seizure locus.J Neurosci. 1997 Feb 1;17(3):875-81. doi: 10.1523/JNEUROSCI.17-03-00875.1997. J Neurosci. 1997. PMID: 8994042 Free PMC article.
-
New missense mutation (G626V) in the predicted selectivity filter of the HERG channel associated with familial long QT syndrome.Hum Mutat. 2000 Jun;15(6):584. doi: 10.1002/1098-1004(200006)15:6<584::AID-HUMU27>3.0.CO;2-L. Hum Mutat. 2000. PMID: 10862104 No abstract available.
-
Is restoration of intracellular trafficking clinically feasible in the long QT syndrome?: The example of HERG mutations.J Cardiovasc Electrophysiol. 2003 Mar;14(3):320-2. doi: 10.1046/j.1540-8167.2003.02363.x. J Cardiovasc Electrophysiol. 2003. PMID: 12716119 Review. No abstract available.
-
A plethora of mechanisms in the HERG-related long QT syndrome. Genetics meets electrophysiology.Cardiovasc Res. 1999 Nov;44(2):242-6. doi: 10.1016/s0008-6363(99)00224-2. Cardiovasc Res. 1999. PMID: 10690299 Review. No abstract available.
Cited by
-
Investigating rare and ultrarare epilepsy syndromes with Drosophila models.Fac Rev. 2021 Jan 29;10:10. doi: 10.12703/r/10-10. eCollection 2021. Fac Rev. 2021. PMID: 33659928 Free PMC article. Review.
-
Interactions between charged residues in the transmembrane segments of the voltage-sensing domain in the hERG channel.J Membr Biol. 2005 Oct;207(3):169-81. doi: 10.1007/s00232-005-0812-1. J Membr Biol. 2005. PMID: 16550488
-
The environmental toxicant ziram enhances neurotransmitter release and increases neuronal excitability via the EAG family of potassium channels.Neurobiol Dis. 2020 Sep;143:104977. doi: 10.1016/j.nbd.2020.104977. Epub 2020 Jun 16. Neurobiol Dis. 2020. PMID: 32553709 Free PMC article.
-
Probing the outer mouth structure of the HERG channel with peptide toxin footprinting and molecular modeling.Biophys J. 2007 May 15;92(10):3524-40. doi: 10.1529/biophysj.106.097360. Epub 2007 Feb 9. Biophys J. 2007. PMID: 17293393 Free PMC article.
-
ERG3 potassium channel-mediated suppression of neuronal intrinsic excitability and prevention of seizure generation in mice.J Physiol. 2018 Oct;596(19):4729-4752. doi: 10.1113/JP275970. Epub 2018 Sep 7. J Physiol. 2018. PMID: 30016551 Free PMC article.
References
-
- Brodie C, Brody M, Sampson SR. Characterization of the relation between sodium channels and electrical activity in cultured rat skeletal myotubes: regulatory aspects. Brain Res. 1989;488:186–194. - PubMed
-
- Catterall WA. Cellular and molecular biology of voltage-gated sodium channels. Physiol Rev. 1992;72:s15–s48. - PubMed
-
- Chandy KG, Gutman GA. Voltage-gated potassium channel genes. In: North RA, editor. Ligand- and voltage-gated ion channels. CRC; Boca Raton, FL: 1995. pp. 1–71.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
