Methionine residues as endogenous antioxidants in proteins
- PMID: 8986759
- PMCID: PMC26351
- DOI: 10.1073/pnas.93.26.15036
Methionine residues as endogenous antioxidants in proteins
Abstract
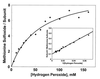
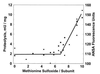
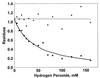
Cysteine and methionine are the two sulfur-containing residues normally found in proteins. Cysteine residues function in the catalytic cycle of many enzymes, and they can form disulfide bonds that contribute to protein structure. In contrast, the specific functions of methionine residues are not known. We propose that methionine residues constitute an important antioxidant defense mechanism. A variety of oxidants react readily with methionine to form methionine sulfoxide, and surface exposed methionine residues create an extremely high concentration of reactant, available as an efficient oxidant scavenger. Reduction back to methionine by methionine sulfoxide reductases would allow the antioxidant system to function catalytically. The effect of hydrogen peroxide exposure upon glutamine synthetase from Escherichia coli was studied as an in vitro model system. Eight of the 16 methionine residues could be oxidized with little effect on catalytic activity of the enzyme. The oxidizable methionine residues were found to be relatively surface exposed, whereas the intact residues were generally buried within the core of the protein. Furthermore, the susceptible residues were physically arranged in an array that guarded the entrance to the active site.
Figures
Similar articles
-
Methionine residues may protect proteins from critical oxidative damage.Mech Ageing Dev. 1999 Mar 15;107(3):323-32. doi: 10.1016/s0047-6374(98)00152-3. Mech Ageing Dev. 1999. PMID: 10360685
-
Oxidative modification of Escherichia coli glutamine synthetase. Decreases in the thermodynamic stability of protein structure and specific changes in the active site conformation.J Biol Chem. 1992 Jan 25;267(3):1872-80. J Biol Chem. 1992. PMID: 1346137
-
Methionine in proteins defends against oxidative stress.FASEB J. 2009 Feb;23(2):464-72. doi: 10.1096/fj.08-118414. Epub 2008 Oct 9. FASEB J. 2009. PMID: 18845767 Free PMC article.
-
Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation.IUBMB Life. 2000 Oct-Nov;50(4-5):301-7. doi: 10.1080/713803735. IUBMB Life. 2000. PMID: 11327324 Review.
-
Methionine in Proteins: It's Not Just for Protein Initiation Anymore.Neurochem Res. 2019 Jan;44(1):247-257. doi: 10.1007/s11064-017-2460-0. Epub 2018 Jan 11. Neurochem Res. 2019. PMID: 29327308 Free PMC article. Review.
Cited by
-
Mass spectrometry-based proteomics as a tool to identify biological matrices in forensic science.Int J Legal Med. 2013 Mar;127(2):287-98. doi: 10.1007/s00414-012-0747-x. Epub 2012 Jul 29. Int J Legal Med. 2013. PMID: 22843116 Free PMC article.
-
The Beneficial Effects of Antioxidants in Health And Diseases.Chronic Obstr Pulm Dis. 2020 Jul;7(3):182-202. doi: 10.15326/jcopdf.7.3.2019.0152. Chronic Obstr Pulm Dis. 2020. PMID: 32558487 Free PMC article. Review.
-
Characterization of Carotenoid-protein Complexes and Gene Expression Analysis Associated with Carotenoid Sequestration in Pigmented Cassava (Manihot Esculenta Crantz) Storage Root.Open Biochem J. 2012;6:116-30. doi: 10.2174/1874091X01206010116. Epub 2012 Nov 26. Open Biochem J. 2012. PMID: 23230451 Free PMC article.
-
Methionine oxidation perturbs the structural core of the prion protein and suggests a generic misfolding pathway.J Biol Chem. 2012 Aug 17;287(34):28263-75. doi: 10.1074/jbc.M112.354779. Epub 2012 May 31. J Biol Chem. 2012. PMID: 22654104 Free PMC article.
-
Drosophila methionine sulfoxide reductase A (MSRA) lacks methionine oxidase activity.Free Radic Biol Med. 2019 Feb 1;131:154-161. doi: 10.1016/j.freeradbiomed.2018.12.001. Epub 2018 Dec 4. Free Radic Biol Med. 2019. PMID: 30529269 Free PMC article.
References
-
- Johnson D, Travis J. J Biol Chem. 1979;254:4022–4026. - PubMed
-
- Rosenberg S, Barr P J, Najarian R C, Hallewell R A. Nature (London) 1994;312:77–80. - PubMed
-
- Stadtman E R. Annu Rev Biochem. 1993;62:797–821. - PubMed
-
- Brot N, Weissbach H. Arch Biochem Biophys. 1983;223:271–281. - PubMed
-
- Shechter Y, Burstein Y, Patchornik A. Biochemistry. 1975;14:4497–4503. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

