Chaperone activity and structure of monomeric polypeptide binding domains of GroEL
- PMID: 8986757
- PMCID: PMC26349
- DOI: 10.1073/pnas.93.26.15024
Chaperone activity and structure of monomeric polypeptide binding domains of GroEL
Abstract
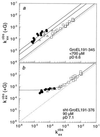
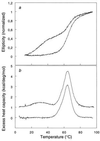
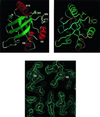
The chaperonin GroEL is a large complex composed of 14 identical 57-kDa subunits that requires ATP and GroES for some of its activities. We find that a monomeric polypeptide corresponding to residues 191 to 345 has the activity of the tetradecamer both in facilitating the refolding of rhodanese and cyclophilin A in the absence of ATP and in catalyzing the unfolding of native barnase. Its crystal structure, solved at 2.5 A resolution, shows a well-ordered domain with the same fold as in intact GroEL. We have thus isolated the active site of the complex allosteric molecular chaperone, which functions as a "minichaperone." This has mechanistic implications: the presence of a central cavity in the GroEL complex is not essential for those representative activities in vitro, and neither are the allosteric properties. The function of the allosteric behavior on the binding of GroES and ATP must be to regulate the affinity of the protein for its various substrates in vivo, where the cavity may also be required for special functions.
Figures


Comment in
-
Dissecting intrinsic chaperonin activity.Proc Natl Acad Sci U S A. 1997 Jan 7;94(1):7-8. doi: 10.1073/pnas.94.1.7. Proc Natl Acad Sci U S A. 1997. PMID: 8990150 Free PMC article. No abstract available.
Similar articles
-
NMR analysis of the binding of a rhodanese peptide to a minichaperone in solution.J Mol Biol. 1999 Sep 10;292(1):181-90. doi: 10.1006/jmbi.1999.3042. J Mol Biol. 1999. PMID: 10493867
-
Destabilization of the complete protein secondary structure on binding to the chaperone GroEL.Nature. 1994 Mar 17;368(6468):261-5. doi: 10.1038/368261a0. Nature. 1994. PMID: 7908413
-
Multiple cycles of global unfolding of GroEL-bound cyclophilin A evidenced by NMR.J Mol Biol. 1997 Sep 5;271(5):803-18. doi: 10.1006/jmbi.1997.1192. J Mol Biol. 1997. PMID: 9299328
-
Molecular chaperone GroEL/ES: unfolding and refolding processes.Biochemistry (Mosc). 2013 Dec;78(13):1405-14. doi: 10.1134/S0006297913130038. Biochemistry (Mosc). 2013. PMID: 24490731 Review.
-
Structural aspects of GroEL function.Curr Opin Struct Biol. 1998 Feb;8(1):93-100. doi: 10.1016/s0959-440x(98)80015-8. Curr Opin Struct Biol. 1998. PMID: 9519301 Review.
Cited by
-
Structural insight into the cooperation of chloroplast chaperonin subunits.BMC Biol. 2016 Apr 12;14:29. doi: 10.1186/s12915-016-0251-8. BMC Biol. 2016. PMID: 27072913 Free PMC article.
-
The inner cavity of Escherichia coli DegP protein is not essential for molecular chaperone and proteolytic activity.J Bacteriol. 2007 Feb;189(3):706-16. doi: 10.1128/JB.01334-06. Epub 2006 Nov 22. J Bacteriol. 2007. PMID: 17122339 Free PMC article.
-
Binding of a burst-phase intermediate formed in the folding of denatured D-glyceraldehyde-3-phosphate dehydrogenase by chaperonin 60 and 8-anilino-1-naphthalenesulphonic acid.Biochem J. 1998 Apr 15;331 ( Pt 2)(Pt 2):505-11. doi: 10.1042/bj3310505. Biochem J. 1998. PMID: 9531491 Free PMC article.
-
Single amino acid substitutions on the surface of Escherichia coli maltose-binding protein can have a profound impact on the solubility of fusion proteins.Protein Sci. 2001 Mar;10(3):622-30. doi: 10.1110/ps.45201. Protein Sci. 2001. PMID: 11344330 Free PMC article.
-
GroEL-assisted protein folding: does it occur within the chaperonin inner cavity?Int J Mol Sci. 2009 May 12;10(5):2066-2083. doi: 10.3390/ijms10052066. Int J Mol Sci. 2009. PMID: 19564940 Free PMC article. Review.
References
-
- Ellis R J, Hartl F U. FASEB J. 1996;10:20–26. - PubMed
-
- Braig K, Otwinowski Z, Hegde R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. - PubMed
-
- Braig K, Adams P D, Brünger A T. Nat Struct Biol. 1995;2:1083–1094. - PubMed
-
- Fenton W A, Kashi Y, Furtak K, Horwich A L. Nature (London) 1994;371:614–619. - PubMed
-
- Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Nature (London) 1994;371:261–264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

