Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans
- PMID: 8917571
- PMCID: PMC24073
- DOI: 10.1073/pnas.93.23.13217
Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans
Abstract
The CST20 gene of Candida albicans was cloned by functional complementation of a deletion of the STE20 gene in Saccharomyces cerevisiae. CST20 encodes a homolog of the Ste20p/p65PAK family of protein kinases. Colonies of C. albicans cells deleted for CST20 revealed defects in the lateral formation of mycelia on synthetic solid "Spider" media. However, hyphal development was not impaired in some other media. A similar phenotype was caused by deletion of HST7, encoding a functional homolog of the S. cerevisiae Ste7p protein kinase. Overexpression of HST7 partially complemented the deletion of CST20. Cells deleted for CST20 were less virulent in a mouse model for systemic candidiasis. Our results suggest that more than one signaling pathway can trigger hyphal development in C. albicans, one of which has a protein kinase cascade that is analogous to the mating response pathway in S. cerevisiae and might have become adapted to the control of mycelial formation in asexual C. albicans.
Figures

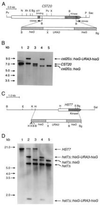

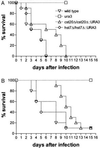

Similar articles
-
Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p.Curr Biol. 1997 Aug 1;7(8):539-46. doi: 10.1016/s0960-9822(06)00252-1. Curr Biol. 1997. PMID: 9259554
-
Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis.Infect Immun. 1998 Jun;66(6):2713-21. doi: 10.1128/IAI.66.6.2713-2721.1998. Infect Immun. 1998. PMID: 9596738 Free PMC article.
-
Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development.Proc Natl Acad Sci U S A. 1996 Nov 12;93(23):13223-8. doi: 10.1073/pnas.93.23.13223. Proc Natl Acad Sci U S A. 1996. PMID: 8917572 Free PMC article.
-
Histidine kinase, two-component signal transduction proteins of Candida albicans and the pathogenesis of candidosis.Mycoses. 1999;42 Suppl 2:49-53. Mycoses. 1999. PMID: 10865904 Review.
-
[Isolation and molecular characterization of the CaPHO85 gene: a negative regulator of phosphate metabolism (PHO system) in Candida albicans].Nihon Ishinkin Gakkai Zasshi. 2003;44(2):101-5. doi: 10.3314/jjmm.44.101. Nihon Ishinkin Gakkai Zasshi. 2003. PMID: 12748591 Review. Japanese.
Cited by
-
The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans.Eukaryot Cell. 2006 Feb;5(2):347-58. doi: 10.1128/EC.5.2.347-358.2006. Eukaryot Cell. 2006. PMID: 16467475 Free PMC article.
-
Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans.Mol Cell Biol. 2000 Dec;20(23):8696-708. doi: 10.1128/MCB.20.23.8696-8708.2000. Mol Cell Biol. 2000. PMID: 11073971 Free PMC article.
-
Self-regulation of Candida albicans population size during GI colonization.PLoS Pathog. 2007 Dec;3(12):e184. doi: 10.1371/journal.ppat.0030184. PLoS Pathog. 2007. PMID: 18069889 Free PMC article.
-
Isolation and characterization of YlBEM1, a gene required for cell polarization and differentiation in the dimorphic yeast Yarrowia lipolytica.Eukaryot Cell. 2002 Aug;1(4):526-37. doi: 10.1128/EC.1.4.526-537.2002. Eukaryot Cell. 2002. PMID: 12456001 Free PMC article.
-
Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans.Eukaryot Cell. 2004 Jun;3(3):724-34. doi: 10.1128/EC.3.3.724-734.2004. Eukaryot Cell. 2004. PMID: 15189993 Free PMC article.
References
-
- Fidel P L, Sobel J D. Trends Microbiol. 1994;2:202–205. - PubMed
-
- Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. - PubMed
-
- Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. - PubMed
-
- Liu H, Styles C, Fink G R. Science. 1993;262:1741–1744. - PubMed
-
- Clark K L, Feldmann P J F, Dignard D, Larocque R, Brown A J P, Lee M G, Thomas D Y, Whiteway M. Mol Gen Genet. 1995;249:609–621. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

