Structural analysis of substrate binding by the molecular chaperone DnaK
- PMID: 8658133
- PMCID: PMC5629921
- DOI: 10.1126/science.272.5268.1606
Structural analysis of substrate binding by the molecular chaperone DnaK
Abstract
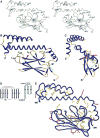
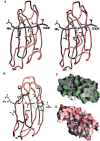
DnaK and other members of the 70-kilodalton heat-shock protein (hsp70) family promote protein folding, interaction, and translocation, both constitutively and in response to stress, by binding to unfolded polypeptide segments. These proteins have two functional units: a substrate-binding portion binds the polypeptide, and an adenosine triphosphatase portion facilitates substrate exchange. The crystal structure of a peptide complex with the substrate-binding unit of DnaK has now been determined at 2.0 angstroms resolution. The structure consists of a beta-sandwich subdomain followed by alpha-helical segments. The peptide is bound to DnaK in an extended conformation through a channel defined by loops from the beta sandwich. An alpha-helical domain stabilizes the complex, but does not contact the peptide directly. This domain is rotated in the molecules of a second crystal lattice, which suggests a model of conformation-dependent substrate binding that features a latch mechanism for maintaining long lifetime complexes.
Figures


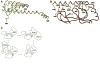
Similar articles
-
Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP.Nat Struct Mol Biol. 2013 Jul;20(7):900-7. doi: 10.1038/nsmb.2583. Epub 2013 May 26. Nat Struct Mol Biol. 2013. PMID: 23708608 Free PMC article.
-
Crystal structure of the molecular chaperone HscA substrate binding domain complexed with the IscU recognition peptide ELPPVKIHC.J Mol Biol. 2004 Sep 24;342(4):1265-78. doi: 10.1016/j.jmb.2004.07.025. J Mol Biol. 2004. PMID: 15351650
-
Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK.J Mol Biol. 2013 Jul 24;425(14):2463-79. doi: 10.1016/j.jmb.2013.03.041. Epub 2013 Apr 2. J Mol Biol. 2013. PMID: 23562829
-
Recent advances in understanding catalysis of protein folding by molecular chaperones.FEBS Lett. 2020 Sep;594(17):2770-2781. doi: 10.1002/1873-3468.13844. Epub 2020 Jun 12. FEBS Lett. 2020. PMID: 32446288 Review.
-
Interaction of Hsp70 chaperones with substrates.Nat Struct Biol. 1997 May;4(5):342-9. doi: 10.1038/nsb0597-342. Nat Struct Biol. 1997. PMID: 9145101 Review.
Cited by
-
Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome.Biomolecules. 2020 Aug 4;10(8):1141. doi: 10.3390/biom10081141. Biomolecules. 2020. PMID: 32759676 Free PMC article. Review.
-
A fluorescent multi-domain protein reveals the unfolding mechanism of Hsp70.Nat Chem Biol. 2023 Feb;19(2):198-205. doi: 10.1038/s41589-022-01162-9. Epub 2022 Oct 20. Nat Chem Biol. 2023. PMID: 36266349 Free PMC article.
-
Theory of Allosteric Regulation in Hsp70 Molecular Chaperones.QRB Discov. 2020;1:e7. doi: 10.1017/qrd.2020.10. Epub 2020 Sep 24. QRB Discov. 2020. PMID: 33738455 Free PMC article.
-
Expression and purification of recombinant proteins based on human prostate stem cell antigen and heat shock protein-70.Exp Ther Med. 2013 Apr;5(4):1161-1164. doi: 10.3892/etm.2013.967. Epub 2013 Feb 20. Exp Ther Med. 2013. PMID: 23596484 Free PMC article.
-
Quantifying the role of chaperones in protein translocation by computational modeling.Front Mol Biosci. 2015 Mar 23;2:8. doi: 10.3389/fmolb.2015.00008. eCollection 2015. Front Mol Biosci. 2015. PMID: 25988176 Free PMC article.
References
-
- Gething MJ, Sambrook J. Nature. 1992;355:33. - PubMed
- Hendrick JP, Hartl FU. Annu Rev Biochem. 1993;62:349. - PubMed
- Morimoto RI, Tissieres A, Georgopoulos C. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto RI, Tissieres A, Georgopoulos C, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994.
-
- Pelham HRB. Cell. 1986;46:959. - PubMed
-
- Rothman JE. ibid. 1989;59:591. - PubMed
-
- Georgopoulos C. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto RI, Tissieres A, Georgopoulos C, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1994. pp. 209–249.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

