Clathrin binding and assembly activities of expressed domains of the synapse-specific clathrin assembly protein AP-3
- PMID: 7738035
- PMCID: PMC4447087
- DOI: 10.1074/jbc.270.18.10933
Clathrin binding and assembly activities of expressed domains of the synapse-specific clathrin assembly protein AP-3
Abstract
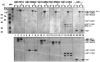
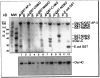
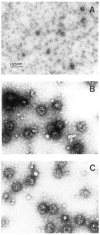
We separately expressed the 58-kDa C-terminal, 42-kDa middle, 16-kDa C-terminal, and 33-kDa N-terminal regions of AP-3 (also called F1-20, AP180, NP185, and pp155), and determined their clathrin binding and assembly properties. The 58-kDa C-terminal region of AP-3 is able to bind to clathrin triskelia and assemble them into a homogeneous population of clathrin cages and will also bind to preassembled clathrin cages. The 42-kDa central region of AP-3 can bind to both clathrin triskelia and to clathrin cages, but cannot assemble clathrin triskelia into clathrin cages. The 16-kDa C-terminal region of AP-3 can bind to clathrin cages, but cannot bind to clathrin triskelia or assemble clathrin triskelia into clathrin cages. The clathrin binding activities of the 42-kDa central region and 16-kDa C-terminal region are weaker than the corresponding activity of either the 58-kDa C-terminal region or full-length AP-3. Previous efforts had mapped a clathrin binding site within the N-terminal 33 kDa of AP-3 (Murphy, J. E., Pleasure, I. T., Puszkin, S., Prasad, K., and Keen, J. H. (1991) J. Biol. Chem. 266, 4401-4408; Morris, S. A., Schroder, S., Plessmann, U., Weber, K., and Ungewickell, E. (1993) EMBO J. 12, 667-675). However, although the N-terminal 33 kDa of AP-3 is able to bind to clathrin triskelia (Murphy, J. E., Pleasure, I. T., Puszkin, S., Prasad, K., and Keen, J. H. (1991) J. Biol. Chem. 266, 4401-4408; Ye, W., and Lafer, E. M. (1995) J. Neurosci. Res. 41, 15-26), it does not promote their assembly into clathrin cages (Murphy, J. E., Pleasure, I. T., Puszkin, S., Prasad, K., and Keen, J. H. (1991) J. Biol. CHem. 266, 4401-4408; Ye, W., and Lafer, E. M. (1995) J. Neurosci. Res. 41, 15-26) or bind to preassembled clathrin cages (Ye, W., and Lafer, E. M. (1995) J. Neurosci. Res. 41, 15-26). It appears that the smallest functional unit that carries out all of the reported clathrin binding and assembly properties of AP-3, essentially as well as the full-length protein, is the 58-kDa C-terminal region.
Figures



Similar articles
-
Bacterially expressed F1-20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission.J Neurosci Res. 1995 May 1;41(1):15-26. doi: 10.1002/jnr.490410104. J Neurosci Res. 1995. PMID: 7674375 Free PMC article.
-
Recognition sites for clathrin-associated proteins AP-2 and AP-3 on clathrin triskelia.J Biol Chem. 1992 May 25;267(15):10850-5. J Biol Chem. 1992. PMID: 1587861
-
Clathrin assembly protein AP-3. The identity of the 155K protein, AP 180, and NP185 and demonstration of a clathrin binding domain.J Biol Chem. 1991 Mar 5;266(7):4401-8. J Biol Chem. 1991. PMID: 1999423
-
Endocytosis: an assembly protein for clathrin cages.Curr Biol. 1999 May 6;9(9):R332-5. doi: 10.1016/s0960-9822(99)80206-1. Curr Biol. 1999. PMID: 10330371 Review.
-
Neuronal protein NP185 is developmentally regulated, initially expressed during synaptogenesis, and localized in synaptic terminals.Mol Neurobiol. 1992 Summer-Fall;6(2-3):253-83. doi: 10.1007/BF02780557. Mol Neurobiol. 1992. PMID: 1476676 Review.
Cited by
-
A role for the clathrin assembly domain of AP180 in synaptic vesicle endocytosis.J Neurosci. 1999 Dec 1;19(23):10201-12. doi: 10.1523/JNEUROSCI.19-23-10201.1999. J Neurosci. 1999. PMID: 10575017 Free PMC article.
-
A Novel Sequence in AP180 and CALM Promotes Efficient Clathrin Binding and Assembly.PLoS One. 2016 Aug 30;11(8):e0162050. doi: 10.1371/journal.pone.0162050. eCollection 2016. PLoS One. 2016. PMID: 27574975 Free PMC article.
-
The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family.Proc Natl Acad Sci U S A. 1996 May 14;93(10):4804-9. doi: 10.1073/pnas.93.10.4804. Proc Natl Acad Sci U S A. 1996. PMID: 8643484 Free PMC article.
-
Dual interaction of synaptotagmin with mu2- and alpha-adaptin facilitates clathrin-coated pit nucleation.EMBO J. 2000 Nov 15;19(22):6011-9. doi: 10.1093/emboj/19.22.6011. EMBO J. 2000. PMID: 11080148 Free PMC article.
-
A conserved clathrin assembly motif essential for synaptic vesicle endocytosis.J Neurosci. 2000 Dec 1;20(23):8667-76. doi: 10.1523/JNEUROSCI.20-23-08667.2000. J Neurosci. 2000. PMID: 11102472 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

