Protein folding intermediates: native-state hydrogen exchange
- PMID: 7618079
- PMCID: PMC3432310
- DOI: 10.1126/science.7618079
Protein folding intermediates: native-state hydrogen exchange
Abstract
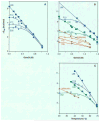
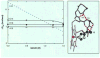
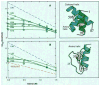
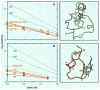
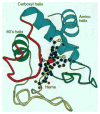
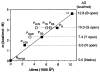
The hydrogen exchange behavior of native cytochrome c in low concentrations of denaturant reveals a sequence of metastable, partially unfolded forms that occupy free energy levels reaching up to the fully unfolded state. The step from one form to another is accomplished by the unfolding of one or more cooperative units of structure. The cooperative units are entire omega loops or mutually stabilizing pairs of whole helices and loops. The partially unfolded forms detected by hydrogen exchange appear to represent the major intermediates in the reversible, dynamic unfolding reactions that occur even at native conditions and thus may define the major pathway for cytochrome c folding.
Figures

Similar articles
-
Relationship between the native-state hydrogen exchange and folding pathways of a four-helix bundle protein.Biochemistry. 2002 Jun 25;41(25):7998-8003. doi: 10.1021/bi025872n. Biochemistry. 2002. PMID: 12069590
-
Kinetic mechanism of folding and unfolding of Rhodobacter capsulatus cytochrome c2.Biochemistry. 1996 Dec 24;35(51):16852-62. doi: 10.1021/bi961976k. Biochemistry. 1996. PMID: 8988024
-
Folding of horse cytochrome c in the reduced state.J Mol Biol. 2001 Oct 5;312(5):1135-60. doi: 10.1006/jmbi.2001.4993. J Mol Biol. 2001. PMID: 11580255
-
Protein folding intermediates and pathways studied by hydrogen exchange.Annu Rev Biophys Biomol Struct. 2000;29:213-38. doi: 10.1146/annurev.biophys.29.1.213. Annu Rev Biophys Biomol Struct. 2000. PMID: 10940248 Review.
-
Future directions in folding: the multi-state nature of protein structure.Proteins. 1996 Feb;24(2):145-51. doi: 10.1002/(SICI)1097-0134(199602)24:2<145::AID-PROT1>3.0.CO;2-I. Proteins. 1996. PMID: 8820481 Review.
Cited by
-
Radiation damping in modern NMR experiments: progress and challenges.Prog Nucl Magn Reson Spectrosc. 2013 Jan;68:41-57. doi: 10.1016/j.pnmrs.2012.06.001. Epub 2012 Jun 15. Prog Nucl Magn Reson Spectrosc. 2013. PMID: 23398972 Free PMC article. No abstract available.
-
Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity.MAbs. 2016 Nov/Dec;8(8):1561-1574. doi: 10.1080/19420862.2016.1222342. Epub 2016 Aug 25. MAbs. 2016. PMID: 27560842 Free PMC article.
-
Urea-Dependent Adenylate Kinase Activation following Redistribution of Structural States.Biophys J. 2016 Oct 4;111(7):1385-1395. doi: 10.1016/j.bpj.2016.08.028. Biophys J. 2016. PMID: 27705762 Free PMC article.
-
Characterization of N-terminal amino group-heme ligation emerging upon guanidine hydrochloric acid induced unfolding of Hydrogenobacter thermophilus ferricytochrome c552.J Biol Inorg Chem. 2008 Jan;13(1):25-34. doi: 10.1007/s00775-007-0298-7. Epub 2007 Sep 22. J Biol Inorg Chem. 2008. PMID: 17899223
-
Apparent tradeoff of higher activity in MMP-12 for enhanced stability and flexibility in MMP-3.Biophys J. 2010 Jul 7;99(1):273-83. doi: 10.1016/j.bpj.2010.04.002. Biophys J. 2010. PMID: 20655856 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

