Codon Bias of the DDR1 Gene and Transcription Factor EHF in Multiple Species
- PMID: 39409024
- PMCID: PMC11477322
- DOI: 10.3390/ijms251910696
Codon Bias of the DDR1 Gene and Transcription Factor EHF in Multiple Species
Abstract
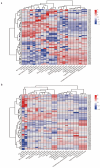
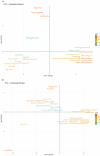
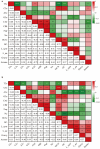
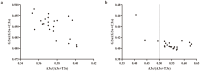
Milk production is an essential economic trait in cattle, and understanding the genetic regulation of this trait can enhance breeding strategies. The discoidin domain receptor 1 (DDR1) gene has been identified as a key candidate gene that influences milk production, and ETS homologous factor (EHF) is recognized as a critical transcription factor that regulates DDR1 expression. Codon usage bias, which affects gene expression and protein function, has not been fully explored in cattle. This study aims to examine the codon usage bias of DDR1 and EHF transcription factors to understand their roles in dairy production traits. Data from 24 species revealed that both DDR1 and EHF predominantly used G/C-ending codons, with the GC3 content averaging 75.49% for DDR1 and 61.72% for EHF. Synonymous codon usage analysis identified high-frequency codons for both DDR1 and EHF, with 17 codons common to both genes. Correlation analysis indicated a negative relationship between the effective number of codons and codon adaptation index for both DDR1 and EHF. Phylogenetic and clustering analyses revealed similar codon usage patterns among closely related species. These findings suggest that EHF plays a crucial role in regulating DDR1 expression, offering new insights into genetically regulating milk production in cattle.
Keywords: DDR1; EHF; cattle; codon usage bias; lactation regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Analysis of codon usage bias of WRKY transcription factors in Helianthus annuus.BMC Genom Data. 2022 Jun 20;23(1):46. doi: 10.1186/s12863-022-01064-8. BMC Genom Data. 2022. PMID: 35725374 Free PMC article.
-
Codon usage characterization and phylogenetic analysis of the mitochondrial genome in Hemerocallis citrina.BMC Genom Data. 2024 Jan 13;25(1):6. doi: 10.1186/s12863-024-01191-4. BMC Genom Data. 2024. PMID: 38218810 Free PMC article.
-
Codon usage bias and dinucleotide preference in 29 Drosophila species.G3 (Bethesda). 2021 Aug 7;11(8):jkab191. doi: 10.1093/g3journal/jkab191. G3 (Bethesda). 2021. PMID: 34849812 Free PMC article.
-
Codon usage bias.Mol Biol Rep. 2022 Jan;49(1):539-565. doi: 10.1007/s11033-021-06749-4. Epub 2021 Nov 25. Mol Biol Rep. 2022. PMID: 34822069 Free PMC article. Review.
-
Codon usage and codon pair patterns in non-grass monocot genomes.Ann Bot. 2017 Nov 28;120(6):893-909. doi: 10.1093/aob/mcx112. Ann Bot. 2017. PMID: 29155926 Free PMC article. Review.
References
-
- Barker K.T., Martindale J.E., Mitchell P.J., Kamalati T., Page M.J., Phippard D.J., Dale T.C., Gusterson B.A., Crompton M.R. Expression patterns of the novel receptor-like tyrosine kinase, DDR, in human breast tumours. Oncogene. 1995;10:569–575. - PubMed
-
- Rauner G., Jin D.X., Miller D.H., Gierahn T.M., Li C.M., Sokol E.S., Feng Y.X., Mathis R.A., Love J.C., Gupta P.B., et al. Breast tissue regeneration is driven by cell-matrix interactions coordinating multi-lineage stem cell differentiation through DDR1. Nat. Commun. 2021;12:7116. doi: 10.1038/s41467-021-27401-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

