GCphase: an SNP phasing method using a graph partition and error correction algorithm
- PMID: 39160480
- PMCID: PMC11331634
- DOI: 10.1186/s12859-024-05901-8
GCphase: an SNP phasing method using a graph partition and error correction algorithm
Abstract
Background: The utilization of long reads for single nucleotide polymorphism (SNP) phasing has become popular, providing substantial support for research on human diseases and genetic studies in animals and plants. However, due to the complexity of the linkage relationships between SNP loci and sequencing errors in the reads, the recent methods still cannot yield satisfactory results.
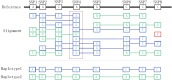
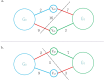
Results: In this study, we present a graph-based algorithm, GCphase, which utilizes the minimum cut algorithm to perform phasing. First, based on alignment between long reads and the reference genome, GCphase filters out ambiguous SNP sites and useless read information. Second, GCphase constructs a graph in which a vertex represents alleles of an SNP locus and each edge represents the presence of read support; moreover, GCphase adopts a graph minimum-cut algorithm to phase the SNPs. Next, GCpahse uses two error correction steps to refine the phasing results obtained from the previous step, effectively reducing the error rate. Finally, GCphase obtains the phase block. GCphase was compared to three other methods, WhatsHap, HapCUT2, and LongPhase, on the Nanopore and PacBio long-read datasets. The code is available from https://github.com/baimawjy/GCphase .
Conclusions: Experimental results show that GCphase under different sequencing depths of different data has the least number of switch errors and the highest accuracy compared with other methods.
Keywords: Error correction; Graph minimum-cut algorithm; Haplotype assembly; SNP phasing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
DCHap: A Divide-and-Conquer Haplotype Phasing Algorithm for Third-Generation Sequences.IEEE/ACM Trans Comput Biol Bioinform. 2022 May-Jun;19(3):1277-1284. doi: 10.1109/TCBB.2020.3005673. Epub 2022 Jun 3. IEEE/ACM Trans Comput Biol Bioinform. 2022. PMID: 32750878
-
HapCUT2: A Method for Phasing Genomes Using Experimental Sequence Data.Methods Mol Biol. 2023;2590:139-147. doi: 10.1007/978-1-0716-2819-5_9. Methods Mol Biol. 2023. PMID: 36335497
-
A hybrid and scalable error correction algorithm for indel and substitution errors of long reads.BMC Genomics. 2019 Dec 20;20(Suppl 11):948. doi: 10.1186/s12864-019-6286-9. BMC Genomics. 2019. PMID: 31856721 Free PMC article.
-
Towards accurate, contiguous and complete alignment-based polyploid phasing algorithms.Genomics. 2022 May;114(3):110369. doi: 10.1016/j.ygeno.2022.110369. Epub 2022 Apr 26. Genomics. 2022. PMID: 35483655 Review.
-
A comprehensive evaluation of long read error correction methods.BMC Genomics. 2020 Dec 21;21(Suppl 6):889. doi: 10.1186/s12864-020-07227-0. BMC Genomics. 2020. PMID: 33349243 Free PMC article. Review.
Cited by
-
Genomic resources, opportunities, and prospects for accelerated improvement of millets.Theor Appl Genet. 2024 Nov 20;137(12):273. doi: 10.1007/s00122-024-04777-9. Theor Appl Genet. 2024. PMID: 39565376 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

