Novel drug resistance mechanisms and drug targets in BRAF-mutated peritoneal metastasis from colorectal cancer
- PMID: 38982444
- PMCID: PMC11234641
- DOI: 10.1186/s12967-024-05467-2
Novel drug resistance mechanisms and drug targets in BRAF-mutated peritoneal metastasis from colorectal cancer
Abstract
Background: Patients with peritoneal metastasis from colorectal cancer (PM-CRC) have inferior prognosis and respond particularly poorly to chemotherapy. This study aims to identify the molecular explanation for the observed clinical behavior and suggest novel treatment strategies in PM-CRC.
Methods: Tumor samples (230) from a Norwegian national cohort undergoing surgery and hyperthermic intraperitoneal chemotherapy (HIPEC) with mitomycin C (MMC) for PM-CRC were subjected to targeted DNA sequencing, and associations with clinical data were analyzed. mRNA sequencing was conducted on a subset of 30 samples to compare gene expression in tumors harboring BRAF or KRAS mutations and wild-type tumors.
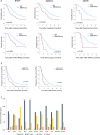
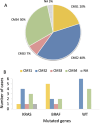
Results: BRAF mutations were detected in 27% of the patients, and the BRAF-mutated subgroup had inferior overall survival compared to wild-type cases (median 16 vs 36 months, respectively, p < 0.001). BRAF mutations were associated with RNF43/RSPO aberrations and low expression of negative Wnt regulators (ligand-dependent Wnt activation). Furthermore, BRAF mutations were associated with gene expression changes in transport solute carrier proteins (specifically SLC7A6) and drug metabolism enzymes (CES1 and CYP3A4) that could influence the efficacy of MMC and irinotecan, respectively. BRAF-mutated tumors additionally exhibited increased expression of members of the novel butyrophilin subfamily of immune checkpoint molecules (BTN1A1 and BTNL9).
Conclusions: BRAF mutations were frequently detected and were associated with particularly poor survival in this cohort, possibly related to ligand-dependent Wnt activation and altered drug transport and metabolism that could confer resistance to MMC and irinotecan. Drugs that target ligand-dependent Wnt activation or the BTN immune checkpoints could represent two novel therapy approaches.
Keywords: Colorectal cancer; Drug resistance; Peritoneal metastasis; Therapeutic targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Impact of KRAS, BRAF and microsatellite instability status after cytoreductive surgery and HIPEC in a national cohort of colorectal peritoneal metastasis patients.Br J Cancer. 2022 Mar;126(5):726-735. doi: 10.1038/s41416-021-01620-6. Epub 2021 Dec 9. Br J Cancer. 2022. PMID: 34887523 Free PMC article.
-
KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer.Br J Cancer. 2009 Aug 18;101(4):715-21. doi: 10.1038/sj.bjc.6605177. Epub 2009 Jul 14. Br J Cancer. 2009. PMID: 19603018 Free PMC article.
-
BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis.Ann Oncol. 2015 Oct;26(10):2092-7. doi: 10.1093/annonc/mdv290. Epub 2015 Jul 7. Ann Oncol. 2015. PMID: 26153495
-
The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis.Acta Oncol. 2014 Jul;53(7):852-64. doi: 10.3109/0284186X.2014.895036. Epub 2014 Mar 25. Acta Oncol. 2014. PMID: 24666267 Review.
-
BRAF-Mutated Colorectal Cancer: What Is the Optimal Strategy for Treatment?Curr Treat Options Oncol. 2017 Feb;18(2):9. doi: 10.1007/s11864-017-0453-5. Curr Treat Options Oncol. 2017. PMID: 28214977 Review.
References
-
- Franko J, et al. Prognosis of patients with peritoneal metastatic colorectal cancer given systemic therapy: an analysis of individual patient data from prospective randomised trials from the Analysis and Research in Cancers of the Digestive System (ARCAD) database. Lancet Oncol. 2016;17(12):1709–1719. doi: 10.1016/S1470-2045(16)30500-9. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

