Rab30 facilitates lipid homeostasis during fasting
- PMID: 38796472
- PMCID: PMC11127972
- DOI: 10.1038/s41467-024-48959-x
Rab30 facilitates lipid homeostasis during fasting
Abstract
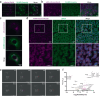
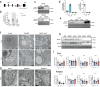
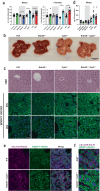
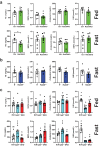
To facilitate inter-tissue communication and the exchange of proteins, lipoproteins, and metabolites with the circulation, hepatocytes have an intricate and efficient intracellular trafficking system regulated by small Rab GTPases. Here, we show that Rab30 is induced in the mouse liver by fasting, which is amplified in liver-specific carnitine palmitoyltransferase 2 knockout mice (Cpt2L-/-) lacking the ability to oxidize fatty acids, in a Pparα-dependent manner. Live-cell super-resolution imaging and in vivo proximity labeling demonstrates that Rab30-marked vesicles are highly dynamic and interact with proteins throughout the secretory pathway. Rab30 whole-body, liver-specific, and Rab30; Cpt2 liver-specific double knockout (DKO) mice are viable with intact Golgi ultrastructure, although Rab30 deficiency in DKO mice suppresses the serum dyslipidemia observed in Cpt2L-/- mice. Corresponding with decreased serum triglyceride and cholesterol levels, DKO mice exhibit decreased circulating but not hepatic ApoA4 protein, indicative of a trafficking defect. Together, these data suggest a role for Rab30 in the selective sorting of lipoproteins to influence hepatocyte and circulating triglyceride levels, particularly during times of excessive lipid burden.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Rab30 is required for the morphological integrity of the Golgi apparatus.Biol Cell. 2012 Feb;104(2):84-101. doi: 10.1111/boc.201100080. Epub 2011 Dec 27. Biol Cell. 2012. PMID: 22188167
-
Golgi-Resident GTPase Rab30 Promotes the Biogenesis of Pathogen-Containing Autophagosomes.PLoS One. 2016 Jan 15;11(1):e0147061. doi: 10.1371/journal.pone.0147061. eCollection 2016. PLoS One. 2016. PMID: 26771875 Free PMC article.
-
Liver-specific deletion of IGF2 mRNA binding protein-2/IMP2 reduces hepatic fatty acid oxidation and increases hepatic triglyceride accumulation.J Biol Chem. 2019 Aug 2;294(31):11944-11951. doi: 10.1074/jbc.RA119.008778. Epub 2019 Jun 17. J Biol Chem. 2019. PMID: 31209109 Free PMC article.
-
mTORC1 activation is not sufficient to suppress hepatic PPARα signaling or ketogenesis.J Biol Chem. 2021 Jul;297(1):100884. doi: 10.1016/j.jbc.2021.100884. Epub 2021 Jun 17. J Biol Chem. 2021. PMID: 34146544 Free PMC article.
-
Discordant hepatic fatty acid oxidation and triglyceride hydrolysis leads to liver disease.JCI Insight. 2021 Jan 25;6(2):e135626. doi: 10.1172/jci.insight.135626. JCI Insight. 2021. PMID: 33491665 Free PMC article.
Cited by
-
Rab2A-mediated Golgi-lipid droplet interactions support very-low-density lipoprotein secretion in hepatocytes.EMBO J. 2024 Dec;43(24):6383-6409. doi: 10.1038/s44318-024-00288-x. Epub 2024 Nov 4. EMBO J. 2024. PMID: 39496977 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

