A Zika virus protein expression screen in Drosophila to investigate targeted host pathways during development
- PMID: 38214058
- PMCID: PMC10924231
- DOI: 10.1242/dmm.050297
A Zika virus protein expression screen in Drosophila to investigate targeted host pathways during development
Abstract
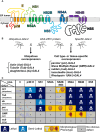
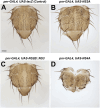
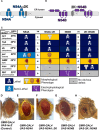
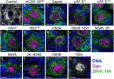
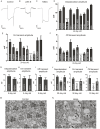
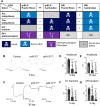
In the past decade, Zika virus (ZIKV) emerged as a global public health concern. Although adult infections are typically mild, maternal infection can lead to adverse fetal outcomes. Understanding how ZIKV proteins disrupt development can provide insights into the molecular mechanisms of disease caused by this virus, which includes microcephaly. In this study, we generated a toolkit to ectopically express ZIKV proteins in vivo in Drosophila melanogaster in a tissue-specific manner using the GAL4/UAS system. We used this toolkit to identify phenotypes and potential host pathways targeted by the virus. Our work identified that expression of most ZIKV proteins caused scorable phenotypes, such as overall lethality, gross morphological defects, reduced brain size and neuronal function defects. We further used this system to identify strain-dependent phenotypes that may have contributed to the increased pathogenesis associated with the outbreak of ZIKV in the Americas in 2015. Our work demonstrates the use of Drosophila as an efficient in vivo model to rapidly decipher how pathogens cause disease and lays the groundwork for further molecular study of ZIKV pathogenesis in flies.
Keywords: Drosophila; Degeneration; Microcephaly; Virus-host targets; Zika virus.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Update of
-
A Zika virus protein expression screen in Drosophila to investigate targeted host pathways during development.bioRxiv [Preprint]. 2023 Apr 29:2023.04.28.538736. doi: 10.1101/2023.04.28.538736. bioRxiv. 2023. Update in: Dis Model Mech. 2024 Feb 1;17(2):dmm050297. doi: 10.1242/dmm.050297 PMID: 37163061 Free PMC article. Updated. Preprint.
Similar articles
-
A Zika virus protein expression screen in Drosophila to investigate targeted host pathways during development.bioRxiv [Preprint]. 2023 Apr 29:2023.04.28.538736. doi: 10.1101/2023.04.28.538736. bioRxiv. 2023. Update in: Dis Model Mech. 2024 Feb 1;17(2):dmm050297. doi: 10.1242/dmm.050297 PMID: 37163061 Free PMC article. Updated. Preprint.
-
Zika Virus Induces Sex-Dependent Metabolic Changes in Drosophila melanogaster to Promote Viral Replication.Front Immunol. 2022 Jun 30;13:903860. doi: 10.3389/fimmu.2022.903860. eCollection 2022. Front Immunol. 2022. PMID: 35844546 Free PMC article.
-
Zika virus non-structural protein NS4A restricts eye growth in Drosophila through regulation of JAK/STAT signaling.Dis Model Mech. 2020 Apr 30;13(4):dmm040816. doi: 10.1242/dmm.040816. Dis Model Mech. 2020. PMID: 32152180 Free PMC article.
-
Zika virus outbreak: a review of neurological complications, diagnosis, and treatment options.J Neurovirol. 2018 Jun;24(3):255-272. doi: 10.1007/s13365-018-0614-8. Epub 2018 Feb 13. J Neurovirol. 2018. PMID: 29441490 Review.
-
Zika virus and microcephaly in Southeast Asia: A cause for concern?J Infect Public Health. 2020 Jan;13(1):11-15. doi: 10.1016/j.jiph.2019.09.012. Epub 2019 Oct 24. J Infect Public Health. 2020. PMID: 31669035 Review.
Cited by
-
Molecular functions of ANKLE2 and its implications in human disease.Dis Model Mech. 2024 Apr 1;17(4):dmm050554. doi: 10.1242/dmm.050554. Epub 2024 May 1. Dis Model Mech. 2024. PMID: 38691001 Free PMC article. Review.
-
Supporting the evolution of infectious disease research.Dis Model Mech. 2024 Sep 1;17(9):dmm052112. doi: 10.1242/dmm.052112. Epub 2024 Oct 1. Dis Model Mech. 2024. PMID: 39352121 Free PMC article.
-
Zika virus tropism and pathogenesis: understanding clinical impacts and transmission dynamics.Virol J. 2024 Oct 29;21(1):271. doi: 10.1186/s12985-024-02547-z. Virol J. 2024. PMID: 39472938 Free PMC article. Review.
References
-
- Aragao, M., Holanda, A. C., Brainer-Lima, A. M., Petribu, N. C. L., Castillo, M., van der Linden, V., Serpa, S. C., Tenorio, A. G., Travassos, P. T. C., Cordeiro, M. T.et al. (2017). Nonmicrocephalic infants with congenital Zika syndrome suspected only after neuroimaging evaluation compared with those with microcephaly at birth and postnatally: how large is the Zika Virus “Iceberg”? AJNR Am. J. Neuroradiol. 38, 1427-1434. 10.3174/ajnr.A5216 - DOI - PMC - PubMed
-
- Bellmann, J., Monette, A., Tripathy, V., Sójka, A., Abo-Rady, M., Janosh, A., Bhatnagar, R., Bickle, M., Mouland, A. J. and Sterneckert, J. (2019). Viral Infections Exacerbate FUS-ALS Phenotypes in iPSC-Derived Spinal Neurons in a Virus Species-Specific Manner. Front Cell Neurosci. 13, 480. 10.3389/fncel.2019.00480 - DOI - PMC - PubMed
-
- Bertolli, J., Attell, J. E., Rose, C., Moore, C. A., Melo, F., Staples, J. E., Kotzky, K., Krishna, N., Satterfield-Nash, A., Pereira, I. O.et al. (2020). Functional outcomes among a cohort of children in northeastern brazil meeting criteria for follow-up of congenital Zika virus infection. Am. J. Trop. Med. Hyg. 102, 955-963. 10.4269/ajtmh.19-0961 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

