LncRNAs-circRNAs as Rising Epigenetic Binary Superstars in Regulating Lipid Metabolic Reprogramming of Cancers
- PMID: 37939296
- PMCID: PMC10767464
- DOI: 10.1002/advs.202303570
LncRNAs-circRNAs as Rising Epigenetic Binary Superstars in Regulating Lipid Metabolic Reprogramming of Cancers
Abstract
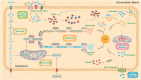
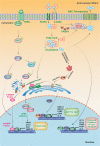
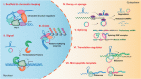
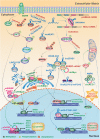
As one of novel hallmarks of cancer, lipid metabolic reprogramming has recently been becoming fascinating and widely studied. Lipid metabolic reprogramming in cancer is shown to support carcinogenesis, progression, distal metastasis, and chemotherapy resistance by generating ATP, biosynthesizing macromolecules, and maintaining appropriate redox status. Notably, increasing evidence confirms that lipid metabolic reprogramming is under the control of dysregulated non-coding RNAs in cancer, especially lncRNAs and circRNAs. This review highlights the present research findings on the aberrantly expressed lncRNAs and circRNAs involved in the lipid metabolic reprogramming of cancer. Emphasis is placed on their regulatory targets in lipid metabolic reprogramming and associated mechanisms, including the clinical relevance in cancer through lipid metabolism modulation. Such insights will be pivotal in identifying new theranostic targets and treatment strategies for cancer patients afflicted with lipid metabolic reprogramming.
Keywords: cancer; cholesterol; circRNAs; fatty acids; lipid metabolic reprogramming; lncRNAs.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Long Noncoding RNAs and Circular RNAs in the Metabolic Reprogramming of Lung Cancer: Functions, Mechanisms, and Clinical Potential.Oxid Med Cell Longev. 2022 Jun 15;2022:4802338. doi: 10.1155/2022/4802338. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35757505 Free PMC article. Review.
-
Long non‑coding RNAs: Key regulators involved in metabolic reprogramming in cancer (Review).Oncol Rep. 2021 May;45(5):54. doi: 10.3892/or.2021.8005. Epub 2021 Mar 24. Oncol Rep. 2021. PMID: 33760177 Review.
-
The emerging regulatory roles of long non-coding RNAs implicated in cancer metabolism.Mol Ther. 2021 Jul 7;29(7):2209-2218. doi: 10.1016/j.ymthe.2021.03.017. Epub 2021 Mar 26. Mol Ther. 2021. PMID: 33775912 Free PMC article. Review.
-
LncRNAs regulate metabolism in cancer.Int J Biol Sci. 2020 Feb 10;16(7):1194-1206. doi: 10.7150/ijbs.40769. eCollection 2020. Int J Biol Sci. 2020. PMID: 32174794 Free PMC article. Review.
-
Amino acid metabolism regulated by lncRNAs: the propellant behind cancer metabolic reprogramming.Cell Commun Signal. 2023 May 1;21(1):87. doi: 10.1186/s12964-023-01116-1. Cell Commun Signal. 2023. PMID: 37127605 Free PMC article. Review.
Cited by
-
Circ_0124346 facilitates cell proliferation of pancreatic adenocarcinoma cells by regulating lipid metabolism via miR-223-3p/ACSL3 axis.Discov Oncol. 2024 Nov 18;15(1):670. doi: 10.1007/s12672-024-01550-8. Discov Oncol. 2024. PMID: 39556281 Free PMC article.
-
Sialylation-associated long non-coding RNA signature predicts the prognosis, tumor microenvironment, and immunotherapy and chemotherapy options in uterine corpus endometrial carcinoma.Cancer Cell Int. 2024 Sep 11;24(1):314. doi: 10.1186/s12935-024-03486-z. Cancer Cell Int. 2024. PMID: 39261877 Free PMC article.
-
Modulating the Expression of Exercise-induced lncRNAs: Implications for Cardiovascular Disease Progression.J Cardiovasc Transl Res. 2024 Jun 10. doi: 10.1007/s12265-024-10530-w. Online ahead of print. J Cardiovasc Transl Res. 2024. PMID: 38858339 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- 2018YFA0106902/National Key Research and Development Program of China
- 82050003/Innovative Program of National Natural Science Foundation of China
- 82371872/National Natural Science Foundation of China
- 32000431/National Natural Science Foundation of China
- 31871297/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials
